Abdulrhman Aljouie
RadGame: An AI-Powered Platform for Radiology Education
Sep 16, 2025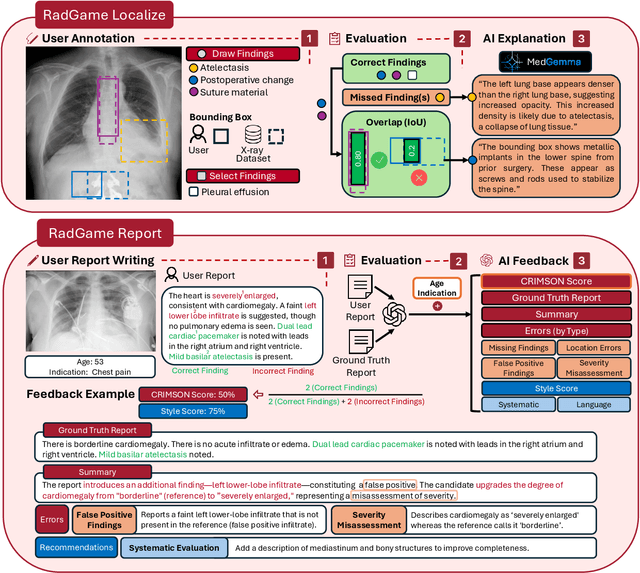
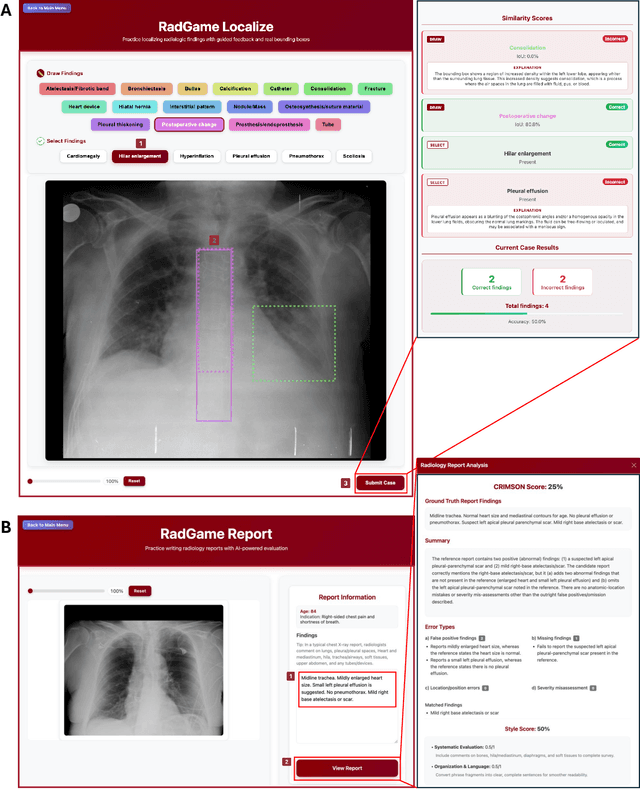
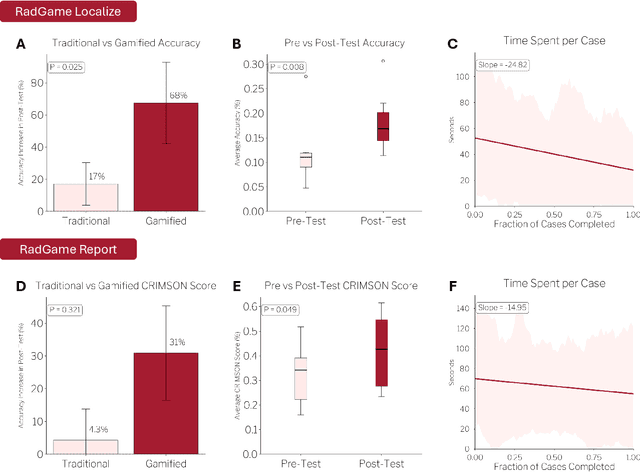
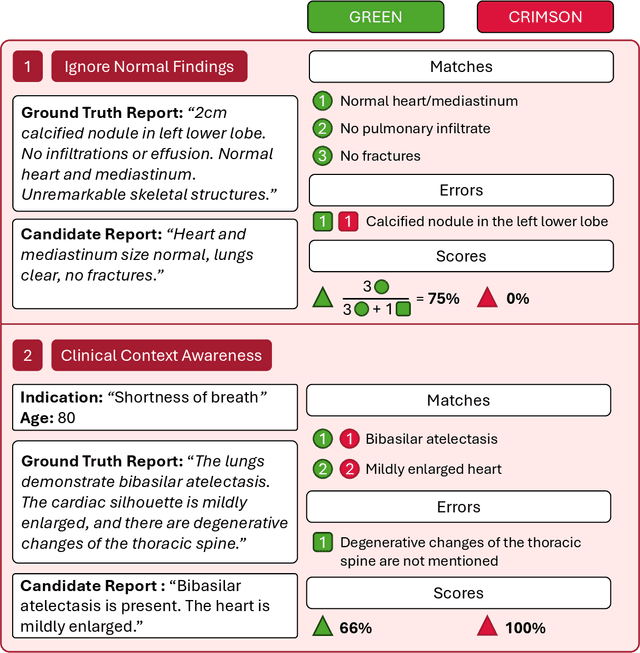
Abstract:We introduce RadGame, an AI-powered gamified platform for radiology education that targets two core skills: localizing findings and generating reports. Traditional radiology training is based on passive exposure to cases or active practice with real-time input from supervising radiologists, limiting opportunities for immediate and scalable feedback. RadGame addresses this gap by combining gamification with large-scale public datasets and automated, AI-driven feedback that provides clear, structured guidance to human learners. In RadGame Localize, players draw bounding boxes around abnormalities, which are automatically compared to radiologist-drawn annotations from public datasets, and visual explanations are generated by vision-language models for user missed findings. In RadGame Report, players compose findings given a chest X-ray, patient age and indication, and receive structured AI feedback based on radiology report generation metrics, highlighting errors and omissions compared to a radiologist's written ground truth report from public datasets, producing a final performance and style score. In a prospective evaluation, participants using RadGame achieved a 68% improvement in localization accuracy compared to 17% with traditional passive methods and a 31% improvement in report-writing accuracy compared to 4% with traditional methods after seeing the same cases. RadGame highlights the potential of AI-driven gamification to deliver scalable, feedback-rich radiology training and reimagines the application of medical AI resources in education.
Towards General Purpose Vision Foundation Models for Medical Image Analysis: An Experimental Study of DINOv2 on Radiology Benchmarks
Dec 07, 2023



Abstract:The integration of deep learning systems into the medical domain has been hindered by the resource-intensive process of data annotation and the inability of these systems to generalize to different data distributions. Foundation models, which are models pre-trained on large datasets, have emerged as a solution to reduce reliance on annotated data and enhance model generalizability and robustness. DINOv2, an open-source foundation model pre-trained with self-supervised learning on 142 million curated natural images, excels in extracting general-purpose visual representations, exhibiting promising capabilities across various vision tasks. Nevertheless, a critical question remains unanswered regarding DINOv2's adaptability to radiological imaging, and the clarity on whether its features are sufficiently general to benefit radiology image analysis is yet to be established. Therefore, this study comprehensively evaluates DINOv2 for radiology, conducting over 100 experiments across diverse modalities (X-ray, CT, and MRI). Tasks include disease classification and organ segmentation on both 2D and 3D images, evaluated under different settings like kNN, few-shot learning, linear-probing, end-to-end fine-tuning, and parameter-efficient fine-tuning, to measure the effectiveness and generalizability of the DINOv2 feature embeddings. Comparative analyses with established medical image analysis models, U-Net and TransUnet for segmentation, and CNN and ViT models pre-trained via supervised, weakly supervised, and self-supervised learning for classification, reveal DINOv2's superior performance in segmentation tasks and competitive results in disease classification. The findings contribute insights to potential avenues for optimizing pre-training strategies for medical imaging and enhancing the broader understanding of DINOv2's role in bridging the gap between natural and radiological image analysis.
HyMNet: a Multimodal Deep Learning System for Hypertension Classification using Fundus Photographs and Cardiometabolic Risk Factors
Oct 02, 2023Abstract:In recent years, deep learning has shown promise in predicting hypertension (HTN) from fundus images. However, most prior research has primarily focused on analyzing a single type of data, which may not capture the full complexity of HTN risk. To address this limitation, this study introduces a multimodal deep learning (MMDL) system, dubbed HyMNet, which combines fundus images and cardiometabolic risk factors, specifically age and gender, to improve hypertension detection capabilities. Our MMDL system uses the DenseNet-201 architecture, pre-trained on ImageNet, for the fundus imaging path and a fully connected neural network for the age and gender path. The two paths are jointly trained by concatenating 64 features output from each path that are then fed into a fusion network. The system was trained on 1,143 retinal images from 626 individuals collected from the Saudi Ministry of National Guard Health Affairs. The results show that the multimodal model that integrates fundus images along with age and gender achieved an AUC of 0.791 [CI: 0.735, 0.848], which outperforms the unimodal model trained solely on fundus photographs that yielded an AUC of 0.766 [CI: 0.705, 0.828] for hypertension detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge