Meng-Chieh Lee
FRAUDGUESS: Spotting and Explaining New Types of Fraud in Million-Scale Financial Data
Sep 19, 2025Abstract:Given a set of financial transactions (who buys from whom, when, and for how much), as well as prior information from buyers and sellers, how can we find fraudulent transactions? If we have labels for some transactions for known types of fraud, we can build a classifier. However, we also want to find new types of fraud, still unknown to the domain experts ('Detection'). Moreover, we also want to provide evidence to experts that supports our opinion ('Justification'). In this paper, we propose FRAUDGUESS, to achieve two goals: (a) for 'Detection', it spots new types of fraud as micro-clusters in a carefully designed feature space; (b) for 'Justification', it uses visualization and heatmaps for evidence, as well as an interactive dashboard for deep dives. FRAUDGUESS is used in real life and is currently considered for deployment in an Anonymous Financial Institution (AFI). Thus, we also present the three new behaviors that FRAUDGUESS discovered in a real, million-scale financial dataset. Two of these behaviors are deemed fraudulent or suspicious by domain experts, catching hundreds of fraudulent transactions that would otherwise go un-noticed.
HybGRAG: Hybrid Retrieval-Augmented Generation on Textual and Relational Knowledge Bases
Dec 20, 2024



Abstract:Given a semi-structured knowledge base (SKB), where text documents are interconnected by relations, how can we effectively retrieve relevant information to answer user questions? Retrieval-Augmented Generation (RAG) retrieves documents to assist large language models (LLMs) in question answering; while Graph RAG (GRAG) uses structured knowledge bases as its knowledge source. However, many questions require both textual and relational information from SKB - referred to as "hybrid" questions - which complicates the retrieval process and underscores the need for a hybrid retrieval method that leverages both information. In this paper, through our empirical analysis, we identify key insights that show why existing methods may struggle with hybrid question answering (HQA) over SKB. Based on these insights, we propose HybGRAG for HQA consisting of a retriever bank and a critic module, with the following advantages: (1) Agentic, it automatically refines the output by incorporating feedback from the critic module, (2) Adaptive, it solves hybrid questions requiring both textual and relational information with the retriever bank, (3) Interpretable, it justifies decision making with intuitive refinement path, and (4) Effective, it surpasses all baselines on HQA benchmarks. In experiments on the STaRK benchmark, HybGRAG achieves significant performance gains, with an average relative improvement in Hit@1 of 51%.
End-To-End Self-tuning Self-supervised Time Series Anomaly Detection
Apr 03, 2024



Abstract:Time series anomaly detection (TSAD) finds many applications such as monitoring environmental sensors, industry KPIs, patient biomarkers, etc. A two-fold challenge for TSAD is a versatile and unsupervised model that can detect various different types of time series anomalies (spikes, discontinuities, trend shifts, etc.) without any labeled data. Modern neural networks have outstanding ability in modeling complex time series. Self-supervised models in particular tackle unsupervised TSAD by transforming the input via various augmentations to create pseudo anomalies for training. However, their performance is sensitive to the choice of augmentation, which is hard to choose in practice, while there exists no effort in the literature on data augmentation tuning for TSAD without labels. Our work aims to fill this gap. We introduce TSAP for TSA "on autoPilot", which can (self-)tune augmentation hyperparameters end-to-end. It stands on two key components: a differentiable augmentation architecture and an unsupervised validation loss to effectively assess the alignment between augmentation type and anomaly type. Case studies show TSAP's ability to effectively select the (discrete) augmentation type and associated (continuous) hyperparameters. In turn, it outperforms established baselines, including SOTA self-supervised models, on diverse TSAD tasks exhibiting different anomaly types.
NetInfoF Framework: Measuring and Exploiting Network Usable Information
Feb 12, 2024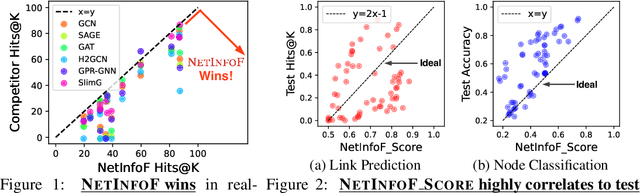
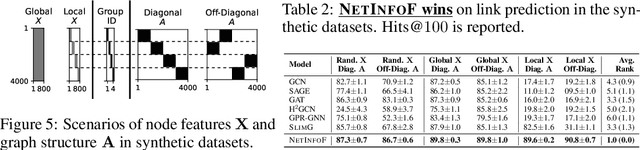
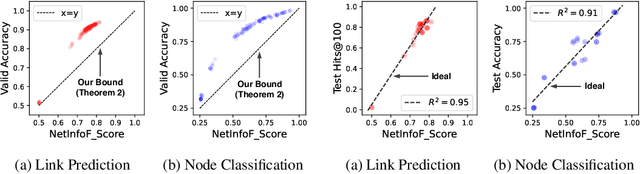

Abstract:Given a node-attributed graph, and a graph task (link prediction or node classification), can we tell if a graph neural network (GNN) will perform well? More specifically, do the graph structure and the node features carry enough usable information for the task? Our goals are (1) to develop a fast tool to measure how much information is in the graph structure and in the node features, and (2) to exploit the information to solve the task, if there is enough. We propose NetInfoF, a framework including NetInfoF_Probe and NetInfoF_Act, for the measurement and the exploitation of network usable information (NUI), respectively. Given a graph data, NetInfoF_Probe measures NUI without any model training, and NetInfoF_Act solves link prediction and node classification, while two modules share the same backbone. In summary, NetInfoF has following notable advantages: (a) General, handling both link prediction and node classification; (b) Principled, with theoretical guarantee and closed-form solution; (c) Effective, thanks to the proposed adjustment to node similarity; (d) Scalable, scaling linearly with the input size. In our carefully designed synthetic datasets, NetInfoF correctly identifies the ground truth of NUI and is the only method being robust to all graph scenarios. Applied on real-world datasets, NetInfoF wins in 11 out of 12 times on link prediction compared to general GNN baselines.
Descriptive Kernel Convolution Network with Improved Random Walk Kernel
Feb 08, 2024



Abstract:Graph kernels used to be the dominant approach to feature engineering for structured data, which are superseded by modern GNNs as the former lacks learnability. Recently, a suite of Kernel Convolution Networks (KCNs) successfully revitalized graph kernels by introducing learnability, which convolves input with learnable hidden graphs using a certain graph kernel. The random walk kernel (RWK) has been used as the default kernel in many KCNs, gaining increasing attention. In this paper, we first revisit the RWK and its current usage in KCNs, revealing several shortcomings of the existing designs, and propose an improved graph kernel RWK+, by introducing color-matching random walks and deriving its efficient computation. We then propose RWK+CN, a KCN that uses RWK+ as the core kernel to learn descriptive graph features with an unsupervised objective, which can not be achieved by GNNs. Further, by unrolling RWK+, we discover its connection with a regular GCN layer, and propose a novel GNN layer RWK+Conv. In the first part of experiments, we demonstrate the descriptive learning ability of RWK+CN with the improved random walk kernel RWK+ on unsupervised pattern mining tasks; in the second part, we show the effectiveness of RWK+ for a variety of KCN architectures and supervised graph learning tasks, and demonstrate the expressiveness of RWK+Conv layer, especially on the graph-level tasks. RWK+ and RWK+Conv adapt to various real-world applications, including web applications such as bot detection in a web-scale Twitter social network, and community classification in Reddit social interaction networks.
UltraProp: Principled and Explainable Propagation on Large Graphs
Dec 31, 2022Abstract:Given a large graph with few node labels, how can we (a) identify the mixed network-effect of the graph and (b) predict the unknown labels accurately and efficiently? This work proposes Network Effect Analysis (NEA) and UltraProp, which are based on two insights: (a) the network-effect (NE) insight: a graph can exhibit not only one of homophily and heterophily, but also both or none in a label-wise manner, and (b) the neighbor-differentiation (ND) insight: neighbors have different degrees of influence on the target node based on the strength of connections. NEA provides a statistical test to check whether a graph exhibits network-effect or not, and surprisingly discovers the absence of NE in many real-world graphs known to have heterophily. UltraProp solves the node classification problem with notable advantages: (a) Accurate, thanks to the network-effect (NE) and neighbor-differentiation (ND) insights; (b) Explainable, precisely estimating the compatibility matrix; (c) Scalable, being linear with the input size and handling graphs with millions of nodes; and (d) Principled, with closed-form formula and theoretical guarantee. Applied on eight real-world graph datasets, UltraProp outperforms top competitors in terms of accuracy and run time, requiring only stock CPU servers. On a large real-world graph with 1.6M nodes and 22.3M edges, UltraProp achieves more than 9 times speedup (12 minutes vs. 2 hours) compared to most competitors.
SlenderGNN: Accurate, Robust, and Interpretable GNN, and the Reasons for its Success
Oct 08, 2022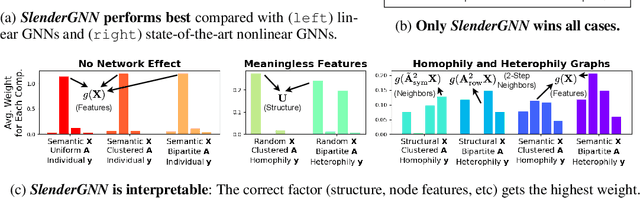
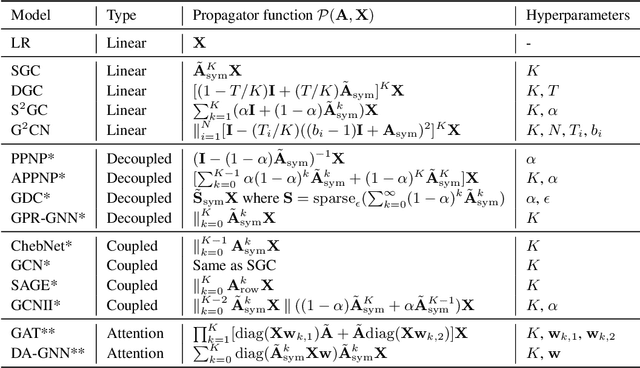
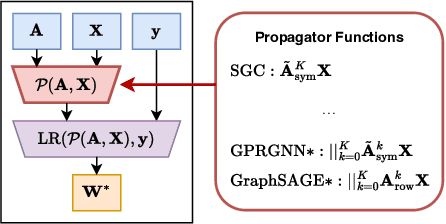
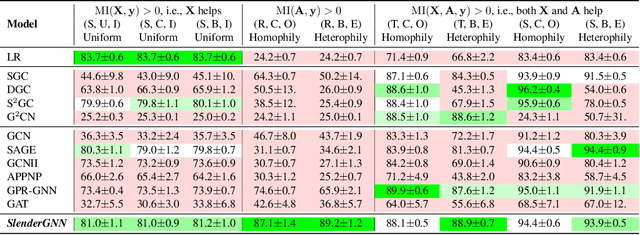
Abstract:Can we design a GNN that is accurate and interpretable at the same time? Could it also be robust to handle the case of homophily, heterophily, or even noisy edges without network effects? We propose SlenderGNN that has all desirable properties: (a) accurate, (b) robust, and (c) interpretable. For the reasons of its success, we had to dig deeper: The result is our GNNLin framework which highlights the fundamental differences among popular GNN models (e.g., feature combination, structural normalization, etc.) and thus reveals the reasons for the success of our SlenderGNN, as well as the reasons for occasional failures of other GNN variants. Thanks to our careful design, SlenderGNN passes all the 'sanity checks' we propose, and it achieves the highest overall accuracy on 9 real-world datasets of both homophily and heterophily graphs, when compared against 10 recent GNN models. Specifically, SlenderGNN exceeds the accuracy of linear GNNs and matches or exceeds the accuracy of nonlinear models with up to 64 times fewer parameters.
gen2Out: Detecting and Ranking Generalized Anomalies
Sep 06, 2021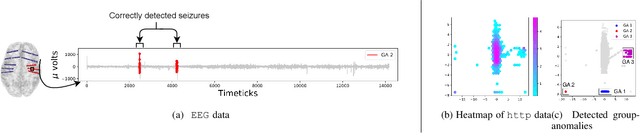
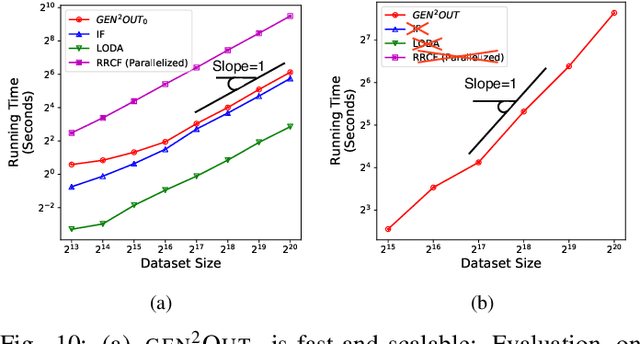
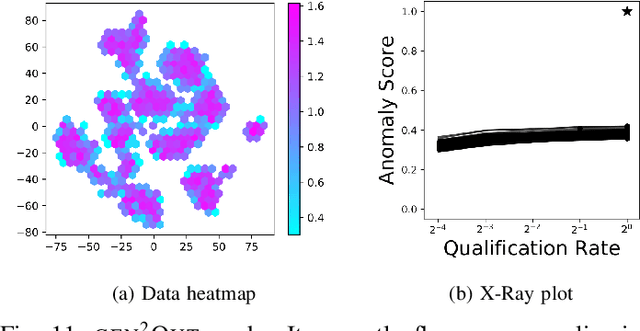
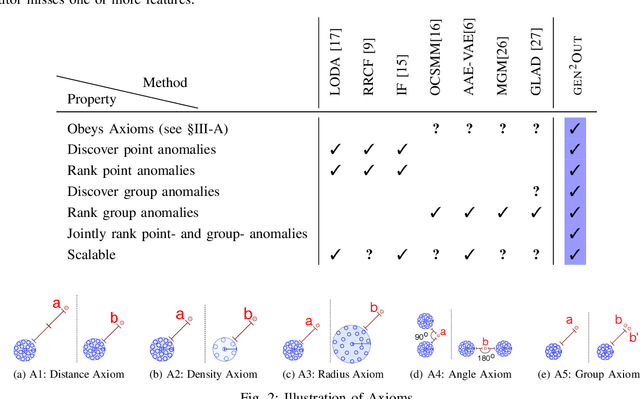
Abstract:In a cloud of m-dimensional data points, how would we spot, as well as rank, both single-point- as well as group- anomalies? We are the first to generalize anomaly detection in two dimensions: The first dimension is that we handle both point-anomalies, as well as group-anomalies, under a unified view -- we shall refer to them as generalized anomalies. The second dimension is that gen2Out not only detects, but also ranks, anomalies in suspiciousness order. Detection, and ranking, of anomalies has numerous applications: For example, in EEG recordings of an epileptic patient, an anomaly may indicate a seizure; in computer network traffic data, it may signify a power failure, or a DoS/DDoS attack. We start by setting some reasonable axioms; surprisingly, none of the earlier methods pass all the axioms. Our main contribution is the gen2Out algorithm, that has the following desirable properties: (a) Principled and Sound anomaly scoring that obeys the axioms for detectors, (b) Doubly-general in that it detects, as well as ranks generalized anomaly -- both point- and group-anomalies, (c) Scalable, it is fast and scalable, linear on input size. (d) Effective, experiments on real-world epileptic recordings (200GB) demonstrate effectiveness of gen2Out as confirmed by clinicians. Experiments on 27 real-world benchmark datasets show that gen2Out detects ground truth groups, matches or outperforms point-anomaly baseline algorithms on accuracy, with no competition for group-anomalies and requires about 2 minutes for 1 million data points on a stock machine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge