Margo Seltzer
Leveraging Predictive Equivalence in Decision Trees
Jun 17, 2025Abstract:Decision trees are widely used for interpretable machine learning due to their clearly structured reasoning process. However, this structure belies a challenge we refer to as predictive equivalence: a given tree's decision boundary can be represented by many different decision trees. The presence of models with identical decision boundaries but different evaluation processes makes model selection challenging. The models will have different variable importance and behave differently in the presence of missing values, but most optimization procedures will arbitrarily choose one such model to return. We present a boolean logical representation of decision trees that does not exhibit predictive equivalence and is faithful to the underlying decision boundary. We apply our representation to several downstream machine learning tasks. Using our representation, we show that decision trees are surprisingly robust to test-time missingness of feature values; we address predictive equivalence's impact on quantifying variable importance; and we present an algorithm to optimize the cost of reaching predictions.
Near Optimal Decision Trees in a SPLIT Second
Feb 21, 2025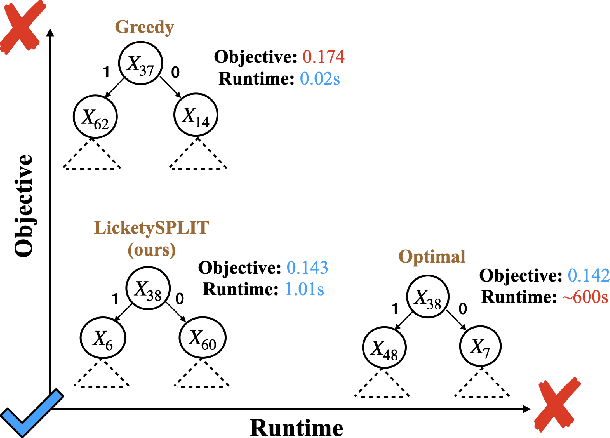

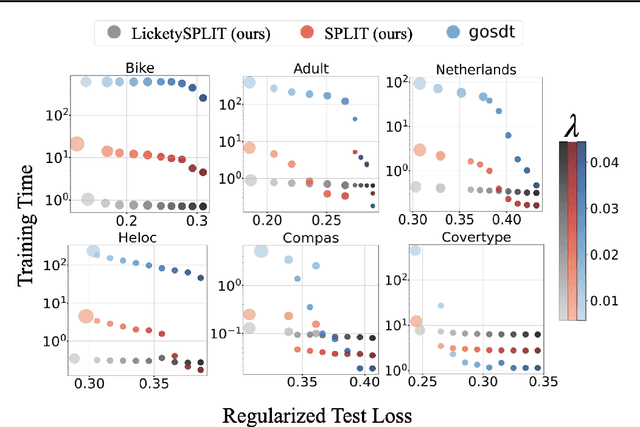

Abstract:Decision tree optimization is fundamental to interpretable machine learning. The most popular approach is to greedily search for the best feature at every decision point, which is fast but provably suboptimal. Recent approaches find the global optimum using branch and bound with dynamic programming, showing substantial improvements in accuracy and sparsity at great cost to scalability. An ideal solution would have the accuracy of an optimal method and the scalability of a greedy method. We introduce a family of algorithms called SPLIT (SParse Lookahead for Interpretable Trees) that moves us significantly forward in achieving this ideal balance. We demonstrate that not all sub-problems need to be solved to optimality to find high quality trees; greediness suffices near the leaves. Since each depth adds an exponential number of possible trees, this change makes our algorithms orders of magnitude faster than existing optimal methods, with negligible loss in performance. We extend this algorithm to allow scalable computation of sets of near-optimal trees (i.e., the Rashomon set).
Interpretable Generalized Additive Models for Datasets with Missing Values
Dec 03, 2024Abstract:Many important datasets contain samples that are missing one or more feature values. Maintaining the interpretability of machine learning models in the presence of such missing data is challenging. Singly or multiply imputing missing values complicates the model's mapping from features to labels. On the other hand, reasoning on indicator variables that represent missingness introduces a potentially large number of additional terms, sacrificing sparsity. We solve these problems with M-GAM, a sparse, generalized, additive modeling approach that incorporates missingness indicators and their interaction terms while maintaining sparsity through l0 regularization. We show that M-GAM provides similar or superior accuracy to prior methods while significantly improving sparsity relative to either imputation or naive inclusion of indicator variables.
HyperBrain: Anomaly Detection for Temporal Hypergraph Brain Networks
Oct 02, 2024


Abstract:Identifying unusual brain activity is a crucial task in neuroscience research, as it aids in the early detection of brain disorders. It is common to represent brain networks as graphs, and researchers have developed various graph-based machine learning methods for analyzing them. However, the majority of existing graph learning tools for the brain face a combination of the following three key limitations. First, they focus only on pairwise correlations between regions of the brain, limiting their ability to capture synchronized activity among larger groups of regions. Second, they model the brain network as a static network, overlooking the temporal changes in the brain. Third, most are designed only for classifying brain networks as healthy or disordered, lacking the ability to identify abnormal brain activity patterns linked to biomarkers associated with disorders. To address these issues, we present HyperBrain, an unsupervised anomaly detection framework for temporal hypergraph brain networks. HyperBrain models fMRI time series data as temporal hypergraphs capturing dynamic higher-order interactions. It then uses a novel customized temporal walk (BrainWalk) and neural encodings to detect abnormal co-activations among brain regions. We evaluate the performance of HyperBrain in both synthetic and real-world settings for Autism Spectrum Disorder and Attention Deficit Hyperactivity Disorder(ADHD). HyperBrain outperforms all other baselines on detecting abnormal co-activations in brain networks. Furthermore, results obtained from HyperBrain are consistent with clinical research on these brain disorders. Our findings suggest that learning temporal and higher-order connections in the brain provides a promising approach to uncover intricate connectivity patterns in brain networks, offering improved diagnosis.
Amazing Things Come From Having Many Good Models
Jul 10, 2024



Abstract:The Rashomon Effect, coined by Leo Breiman, describes the phenomenon that there exist many equally good predictive models for the same dataset. This phenomenon happens for many real datasets and when it does, it sparks both magic and consternation, but mostly magic. In light of the Rashomon Effect, this perspective piece proposes reshaping the way we think about machine learning, particularly for tabular data problems in the nondeterministic (noisy) setting. We address how the Rashomon Effect impacts (1) the existence of simple-yet-accurate models, (2) flexibility to address user preferences, such as fairness and monotonicity, without losing performance, (3) uncertainty in predictions, fairness, and explanations, (4) reliable variable importance, (5) algorithm choice, specifically, providing advanced knowledge of which algorithms might be suitable for a given problem, and (6) public policy. We also discuss a theory of when the Rashomon Effect occurs and why. Our goal is to illustrate how the Rashomon Effect can have a massive impact on the use of machine learning for complex problems in society.
Optimal Sparse Survival Trees
Jan 27, 2024



Abstract:Interpretability is crucial for doctors, hospitals, pharmaceutical companies and biotechnology corporations to analyze and make decisions for high stakes problems that involve human health. Tree-based methods have been widely adopted for \textit{survival analysis} due to their appealing interpretablility and their ability to capture complex relationships. However, most existing methods to produce survival trees rely on heuristic (or greedy) algorithms, which risk producing sub-optimal models. We present a dynamic-programming-with-bounds approach that finds provably-optimal sparse survival tree models, frequently in only a few seconds.
CAT-Walk: Inductive Hypergraph Learning via Set Walks
Jun 19, 2023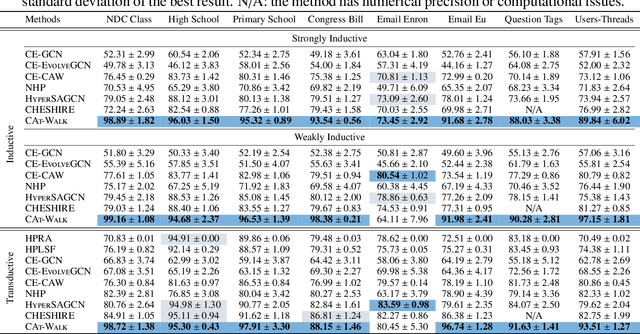
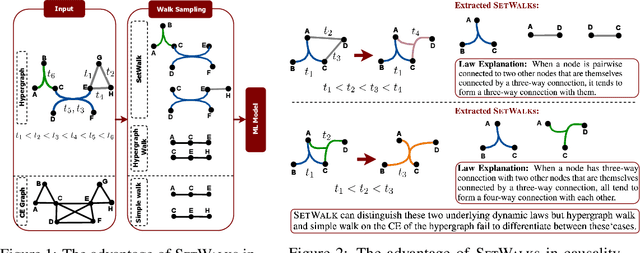
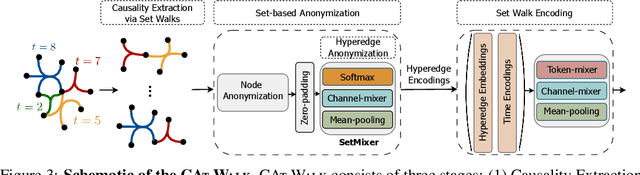
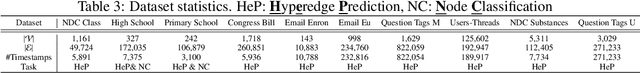
Abstract:Temporal hypergraphs provide a powerful paradigm for modeling time-dependent, higher-order interactions in complex systems. Representation learning for hypergraphs is essential for extracting patterns of the higher-order interactions that are critically important in real-world problems in social network analysis, neuroscience, finance, etc. However, existing methods are typically designed only for specific tasks or static hypergraphs. We present CAT-Walk, an inductive method that learns the underlying dynamic laws that govern the temporal and structural processes underlying a temporal hypergraph. CAT-Walk introduces a temporal, higher-order walk on hypergraphs, SetWalk, that extracts higher-order causal patterns. CAT-Walk uses a novel adaptive and permutation invariant pooling strategy, SetMixer, along with a set-based anonymization process that hides the identity of hyperedges. Finally, we present a simple yet effective neural network model to encode hyperedges. Our evaluation on 10 hypergraph benchmark datasets shows that CAT-Walk attains outstanding performance on temporal hyperedge prediction benchmarks in both inductive and transductive settings. It also shows competitive performance with state-of-the-art methods for node classification.
Understanding and Exploring the Whole Set of Good Sparse Generalized Additive Models
Mar 28, 2023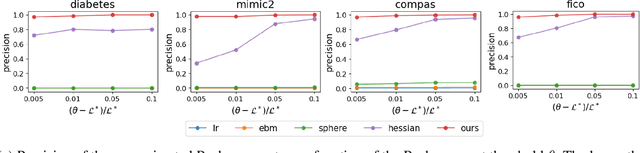
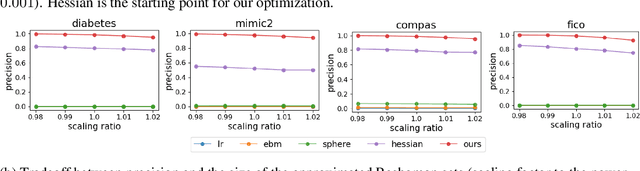
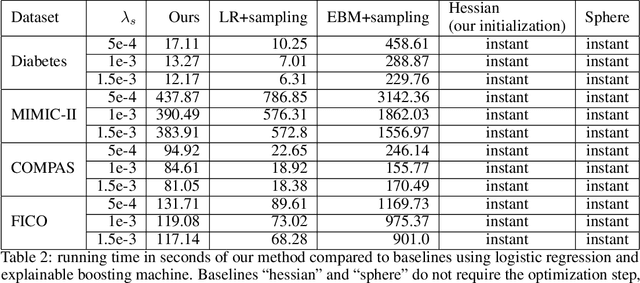
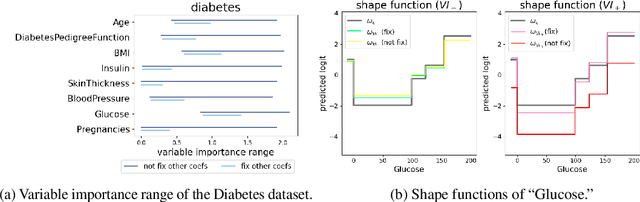
Abstract:In real applications, interaction between machine learning model and domain experts is critical; however, the classical machine learning paradigm that usually produces only a single model does not facilitate such interaction. Approximating and exploring the Rashomon set, i.e., the set of all near-optimal models, addresses this practical challenge by providing the user with a searchable space containing a diverse set of models from which domain experts can choose. We present a technique to efficiently and accurately approximate the Rashomon set of sparse, generalized additive models (GAMs). We present algorithms to approximate the Rashomon set of GAMs with ellipsoids for fixed support sets and use these ellipsoids to approximate Rashomon sets for many different support sets. The approximated Rashomon set serves as a cornerstone to solve practical challenges such as (1) studying the variable importance for the model class; (2) finding models under user-specified constraints (monotonicity, direct editing); (3) investigating sudden changes in the shape functions. Experiments demonstrate the fidelity of the approximated Rashomon set and its effectiveness in solving practical challenges.
Optimal Sparse Regression Trees
Dec 02, 2022



Abstract:Regression trees are one of the oldest forms of AI models, and their predictions can be made without a calculator, which makes them broadly useful, particularly for high-stakes applications. Within the large literature on regression trees, there has been little effort towards full provable optimization, mainly due to the computational hardness of the problem. This work proposes a dynamic-programming-with-bounds approach to the construction of provably-optimal sparse regression trees. We leverage a novel lower bound based on an optimal solution to the k-Means clustering algorithm in 1-dimension over the set of labels. We are often able to find optimal sparse trees in seconds, even for challenging datasets that involve large numbers of samples and highly-correlated features.
Anomaly Detection in Multiplex Dynamic Networks: from Blockchain Security to Brain Disease Prediction
Nov 15, 2022



Abstract:The problem of identifying anomalies in dynamic networks is a fundamental task with a wide range of applications. However, it raises critical challenges due to the complex nature of anomalies, lack of ground truth knowledge, and complex and dynamic interactions in the network. Most existing approaches usually study networks with a single type of connection between vertices, while in many applications interactions between objects vary, yielding multiplex networks. We propose ANOMULY, a general, unsupervised edge anomaly detection framework for multiplex dynamic networks. In each relation type, ANOMULY sees node embeddings at different GNN layers as hierarchical node states and employs a GRU cell to capture temporal properties of the network and update node embeddings over time. We then add an attention mechanism that incorporates information across different types of relations. Our case study on brain networks shows how this approach could be employed as a new tool to understand abnormal brain activity that might reveal a brain disease or disorder. Extensive experiments on nine real-world datasets demonstrate that ANOMULY achieves state-of-the-art performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge