Li Ye
Teeth And Root Canals Segmentation Using ZXYFormer With Uncertainty Guidance And Weight Transfer
Aug 14, 2023



Abstract:This study attempts to segment teeth and root-canals simultaneously from CBCT images, but there are very challenging problems in this process. First, the clinical CBCT image data is very large (e.g., 672 *688 * 688), and the use of downsampling operation will lose useful information about teeth and root canals. Second, teeth and root canals are very different in morphology, and it is difficult for a simple network to identify them precisely. In addition, there are weak edges at the tooth, between tooth and root canal, which makes it very difficult to segment such weak edges. To this end, we propose a coarse-to-fine segmentation method based on inverse feature fusion transformer and uncertainty estimation to address above challenging problems. First, we use the downscaled volume data (e.g., 128 * 128 * 128) to conduct coarse segmentation and map it to the original volume to obtain the area of teeth and root canals. Then, we design a transformer with reverse feature fusion, which can bring better segmentation effect of different morphological objects by transferring deeper features to shallow features. Finally, we design an auxiliary branch to calculate and refine the difficult areas in order to improve the weak edge segmentation performance of teeth and root canals. Through the combined tooth and root canal segmentation experiment of 157 clinical high-resolution CBCT data, it is verified that the proposed method is superior to the existing tooth or root canal segmentation methods.
BERT-based knowledge extraction method of unstructured domain text
Mar 01, 2021Abstract:With the development and business adoption of knowledge graph, there is an increasing demand for extracting entities and relations of knowledge graphs from unstructured domain documents. This makes the automatic knowledge extraction for domain text quite meaningful. This paper proposes a knowledge extraction method based on BERT, which is used to extract knowledge points from unstructured specific domain texts (such as insurance clauses in the insurance industry) automatically to save manpower of knowledge graph construction. Different from the commonly used methods which are based on rules, templates or entity extraction models, this paper converts the domain knowledge points into question and answer pairs and uses the text around the answer in documents as the context. The method adopts a BERT-based model similar to BERT's SQuAD reading comprehension task. The model is fine-tuned. And it is used to directly extract knowledge points from more insurance clauses. According to the test results, the model performance is good.
Combining Offline Causal Inference and Online Bandit Learning for Data Driven Decisions
Jan 16, 2020
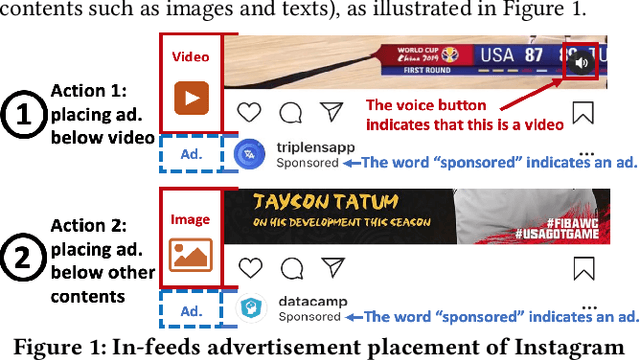
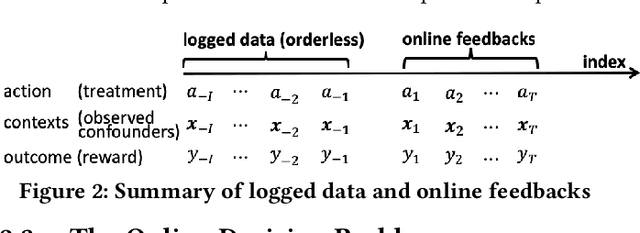
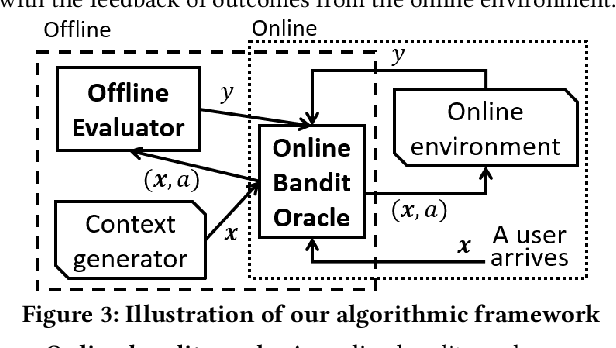
Abstract:A fundamental question for companies is: How to make good decisions with the increasing amount of logged data?. Currently, companies are doing online tests (e.g. A/B tests) before making decisions. However, online tests can be expensive because testing inferior decisions hurt users' experiences. On the other hand, offline causal inference analyzes logged data alone to make decisions, but once a wrong decision is made by the offline causal inference, this wrong decision will continuously to hurt all users' experience. In this paper, we unify offline causal inference and online bandit learning to make the right decision. Our framework is flexible to incorporate various causal inference methods (e.g. matching, weighting) and online bandit methods (e.g. UCB, LinUCB). For these novel combination of algorithms, we derive theoretical bounds on the decision maker's "regret" compared to its optimal decision. We also derive the first regret bound for forest-based online bandit algorithms. Experiments on synthetic data show that our algorithms outperform methods that use only the logged data or only the online feedbacks.
A Large Deformation Diffeomorphic Approach to Registration of CLARITY Images via Mutual Information
Aug 11, 2017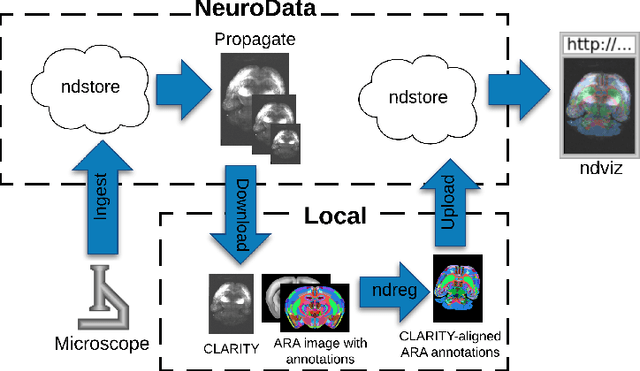
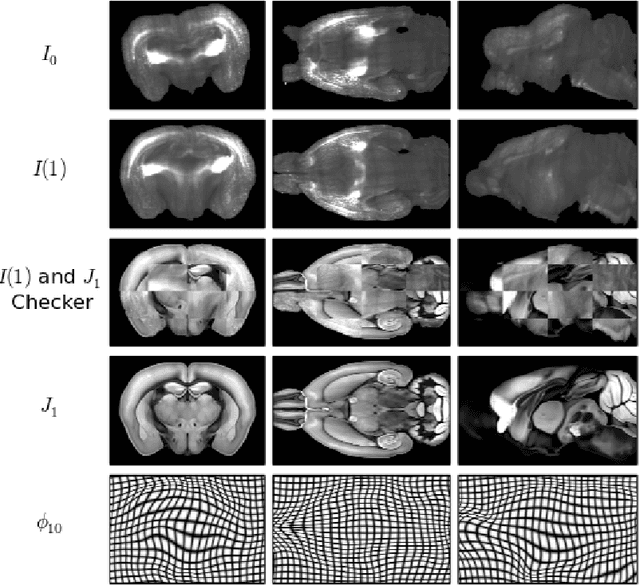
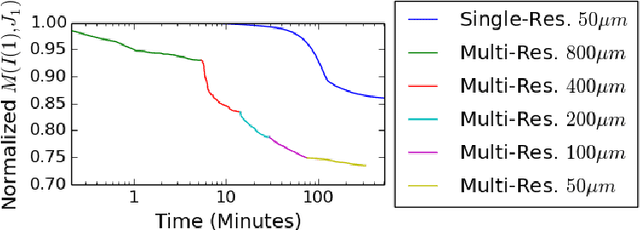
Abstract:CLARITY is a method for converting biological tissues into translucent and porous hydrogel-tissue hybrids. This facilitates interrogation with light sheet microscopy and penetration of molecular probes while avoiding physical slicing. In this work, we develop a pipeline for registering CLARIfied mouse brains to an annotated brain atlas. Due to the novelty of this microscopy technique it is impractical to use absolute intensity values to align these images to existing standard atlases. Thus we adopt a large deformation diffeomorphic approach for registering images via mutual information matching. Furthermore we show how a cascaded multi-resolution approach can improve registration quality while reducing algorithm run time. As acquired image volumes were over a terabyte in size, they were far too large for work on personal computers. Therefore the NeuroData computational infrastructure was deployed for multi-resolution storage and visualization of these images and aligned annotations on the web.
Deformably Registering and Annotating Whole CLARITY Brains to an Atlas via Masked LDDMM
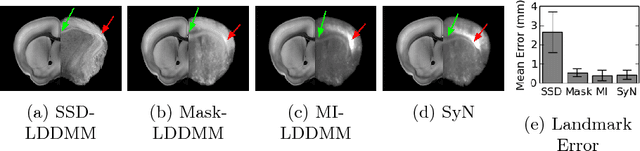
May 06, 2016Abstract:The CLARITY method renders brains optically transparent to enable high-resolution imaging in the structurally intact brain. Anatomically annotating CLARITY brains is necessary for discovering which regions contain signals of interest. Manually annotating whole-brain, terabyte CLARITY images is difficult, time-consuming, subjective, and error-prone. Automatically registering CLARITY images to a pre-annotated brain atlas offers a solution, but is difficult for several reasons. Removal of the brain from the skull and subsequent storage and processing cause variable non-rigid deformations, thus compounding inter-subject anatomical variability. Additionally, the signal in CLARITY images arises from various biochemical contrast agents which only sparsely label brain structures. This sparse labeling challenges the most commonly used registration algorithms that need to match image histogram statistics to the more densely labeled histological brain atlases. The standard method is a multiscale Mutual Information B-spline algorithm that dynamically generates an average template as an intermediate registration target. We determined that this method performs poorly when registering CLARITY brains to the Allen Institute's Mouse Reference Atlas (ARA), because the image histogram statistics are poorly matched. Therefore, we developed a method (Mask-LDDMM) for registering CLARITY images, that automatically find the brain boundary and learns the optimal deformation between the brain and atlas masks. Using Mask-LDDMM without an average template provided better results than the standard approach when registering CLARITY brains to the ARA. The LDDMM pipelines developed here provide a fast automated way to anatomically annotate CLARITY images. Our code is available as open source software at http://NeuroData.io.
Moving Object Detection in Video Using Saliency Map and Subspace Learning
Sep 30, 2015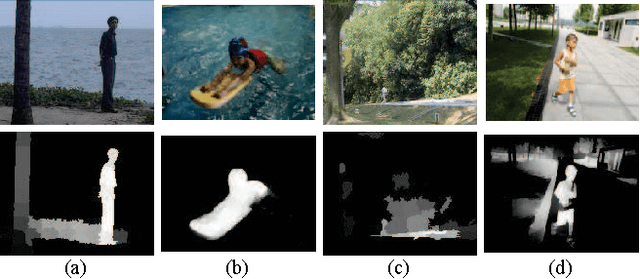

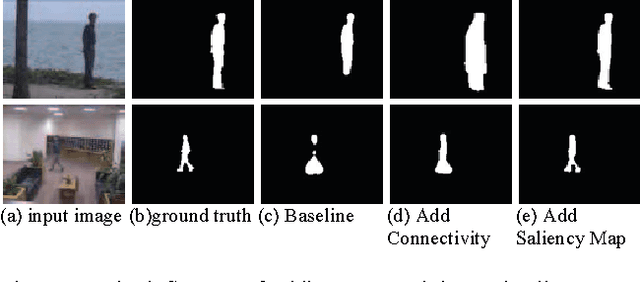
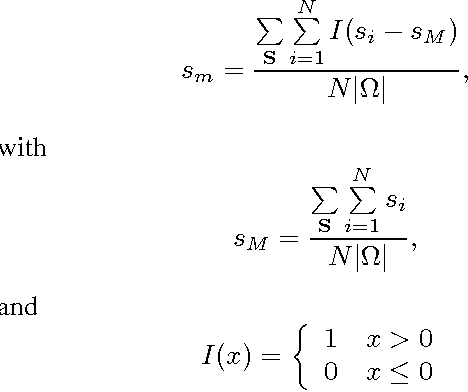
Abstract:Moving object detection is a key to intelligent video analysis. On the one hand, what moves is not only interesting objects but also noise and cluttered background. On the other hand, moving objects without rich texture are prone not to be detected. So there are undesirable false alarms and missed alarms in many algorithms of moving object detection. To reduce the false alarms and missed alarms, in this paper, we propose to incorporate a saliency map into an incremental subspace analysis framework where the saliency map makes estimated background has less chance than foreground (i.e., moving objects) to contain salient objects. The proposed objective function systematically takes account into the properties of sparsity, low-rank, connectivity, and saliency. An alternative minimization algorithm is proposed to seek the optimal solutions. Experimental results on the Perception Test Images Sequences demonstrate that the proposed method is effective in reducing false alarms and missed alarms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge