Alexander D. Baden
A Large Deformation Diffeomorphic Approach to Registration of CLARITY Images via Mutual Information
Aug 11, 2017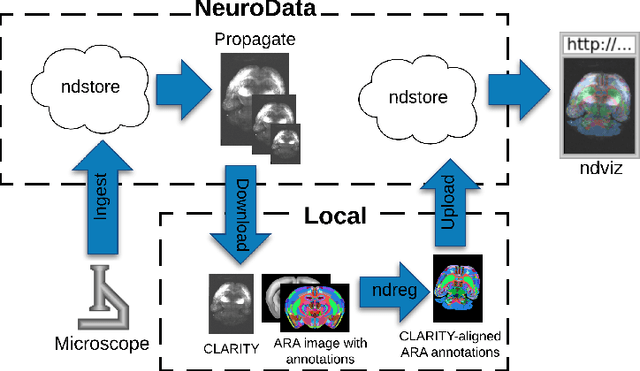
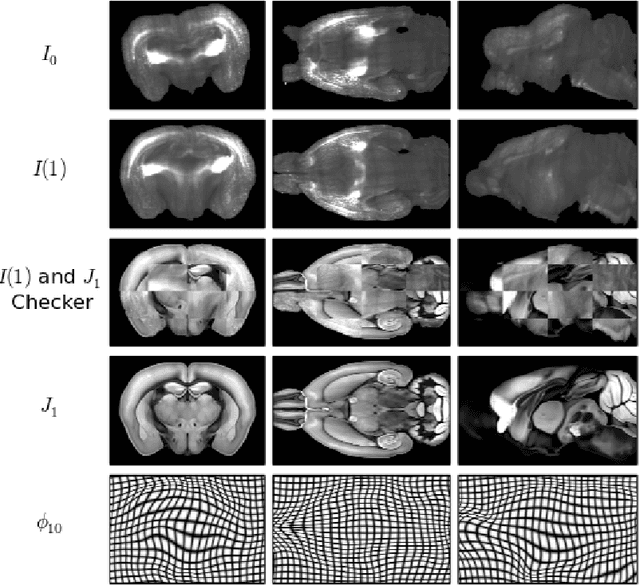
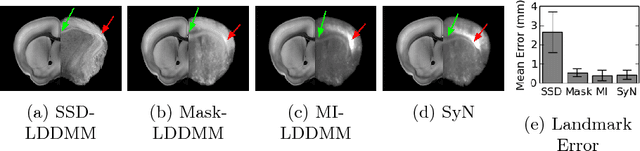
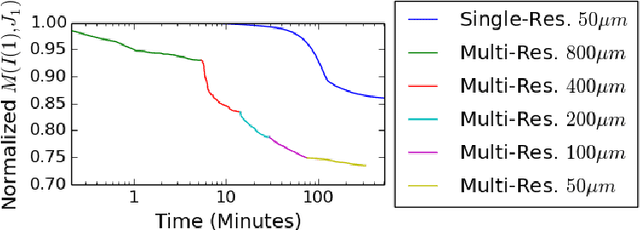
Abstract:CLARITY is a method for converting biological tissues into translucent and porous hydrogel-tissue hybrids. This facilitates interrogation with light sheet microscopy and penetration of molecular probes while avoiding physical slicing. In this work, we develop a pipeline for registering CLARIfied mouse brains to an annotated brain atlas. Due to the novelty of this microscopy technique it is impractical to use absolute intensity values to align these images to existing standard atlases. Thus we adopt a large deformation diffeomorphic approach for registering images via mutual information matching. Furthermore we show how a cascaded multi-resolution approach can improve registration quality while reducing algorithm run time. As acquired image volumes were over a terabyte in size, they were far too large for work on personal computers. Therefore the NeuroData computational infrastructure was deployed for multi-resolution storage and visualization of these images and aligned annotations on the web.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge