Kwang-Hyun Uhm
Multi-Class Segmentation of Aortic Branches and Zones in Computed Tomography Angiography: The AortaSeg24 Challenge
Feb 07, 2025



Abstract:Multi-class segmentation of the aorta in computed tomography angiography (CTA) scans is essential for diagnosing and planning complex endovascular treatments for patients with aortic dissections. However, existing methods reduce aortic segmentation to a binary problem, limiting their ability to measure diameters across different branches and zones. Furthermore, no open-source dataset is currently available to support the development of multi-class aortic segmentation methods. To address this gap, we organized the AortaSeg24 MICCAI Challenge, introducing the first dataset of 100 CTA volumes annotated for 23 clinically relevant aortic branches and zones. This dataset was designed to facilitate both model development and validation. The challenge attracted 121 teams worldwide, with participants leveraging state-of-the-art frameworks such as nnU-Net and exploring novel techniques, including cascaded models, data augmentation strategies, and custom loss functions. We evaluated the submitted algorithms using the Dice Similarity Coefficient (DSC) and Normalized Surface Distance (NSD), highlighting the approaches adopted by the top five performing teams. This paper presents the challenge design, dataset details, evaluation metrics, and an in-depth analysis of the top-performing algorithms. The annotated dataset, evaluation code, and implementations of the leading methods are publicly available to support further research. All resources can be accessed at https://aortaseg24.grand-challenge.org.
Lesion-Aware Cross-Phase Attention Network for Renal Tumor Subtype Classification on Multi-Phase CT Scans
Jun 24, 2024Abstract:Multi-phase computed tomography (CT) has been widely used for the preoperative diagnosis of kidney cancer due to its non-invasive nature and ability to characterize renal lesions. However, since enhancement patterns of renal lesions across CT phases are different even for the same lesion type, the visual assessment by radiologists suffers from inter-observer variability in clinical practice. Although deep learning-based approaches have been recently explored for differential diagnosis of kidney cancer, they do not explicitly model the relationships between CT phases in the network design, limiting the diagnostic performance. In this paper, we propose a novel lesion-aware cross-phase attention network (LACPANet) that can effectively capture temporal dependencies of renal lesions across CT phases to accurately classify the lesions into five major pathological subtypes from time-series multi-phase CT images. We introduce a 3D inter-phase lesion-aware attention mechanism to learn effective 3D lesion features that are used to estimate attention weights describing the inter-phase relations of the enhancement patterns. We also present a multi-scale attention scheme to capture and aggregate temporal patterns of lesion features at different spatial scales for further improvement. Extensive experiments on multi-phase CT scans of kidney cancer patients from the collected dataset demonstrate that our LACPANet outperforms state-of-the-art approaches in diagnostic accuracy.
* This article has been accepted for publication in Computers in Biology and Medicine
Exploring 3D U-Net Training Configurations and Post-Processing Strategies for the MICCAI 2023 Kidney and Tumor Segmentation Challenge
Dec 09, 2023


Abstract:In 2023, it is estimated that 81,800 kidney cancer cases will be newly diagnosed, and 14,890 people will die from this cancer in the United States. Preoperative dynamic contrast-enhanced abdominal computed tomography (CT) is often used for detecting lesions. However, there exists inter-observer variability due to subtle differences in the imaging features of kidney and kidney tumors. In this paper, we explore various 3D U-Net training configurations and effective post-processing strategies for accurate segmentation of kidneys, cysts, and kidney tumors in CT images. We validated our model on the dataset of the 2023 Kidney and Kidney Tumor Segmentation (KiTS23) challenge. Our method took second place in the final ranking of the KiTS23 challenge on unseen test data with an average Dice score of 0.820 and an average Surface Dice of 0.712.
A Unified Multi-Phase CT Synthesis and Classification Framework for Kidney Cancer Diagnosis with Incomplete Data
Dec 09, 2023Abstract:Multi-phase CT is widely adopted for the diagnosis of kidney cancer due to the complementary information among phases. However, the complete set of multi-phase CT is often not available in practical clinical applications. In recent years, there have been some studies to generate the missing modality image from the available data. Nevertheless, the generated images are not guaranteed to be effective for the diagnosis task. In this paper, we propose a unified framework for kidney cancer diagnosis with incomplete multi-phase CT, which simultaneously recovers missing CT images and classifies cancer subtypes using the completed set of images. The advantage of our framework is that it encourages a synthesis model to explicitly learn to generate missing CT phases that are helpful for classifying cancer subtypes. We further incorporate lesion segmentation network into our framework to exploit lesion-level features for effective cancer classification in the whole CT volumes. The proposed framework is based on fully 3D convolutional neural networks to jointly optimize both synthesis and classification of 3D CT volumes. Extensive experiments on both in-house and external datasets demonstrate the effectiveness of our framework for the diagnosis with incomplete data compared with state-of-the-art baselines. In particular, cancer subtype classification using the completed CT data by our method achieves higher performance than the classification using the given incomplete data.
* This article has been accepted for publication in IEEE Journal of Biomedical and Health Informatics
W-Net: Two-stage U-Net with misaligned data for raw-to-RGB mapping
Nov 22, 2019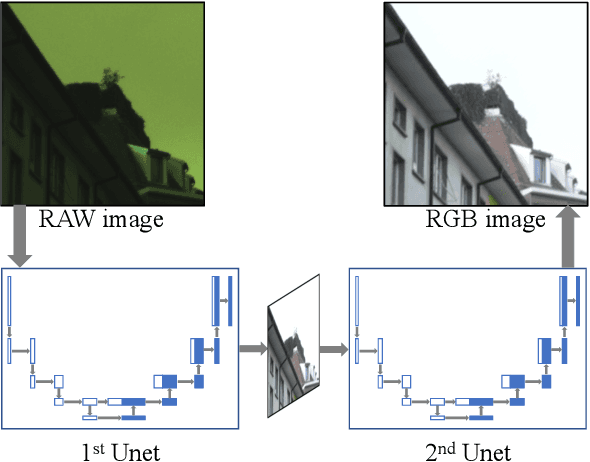
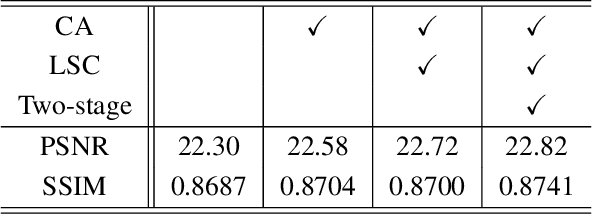
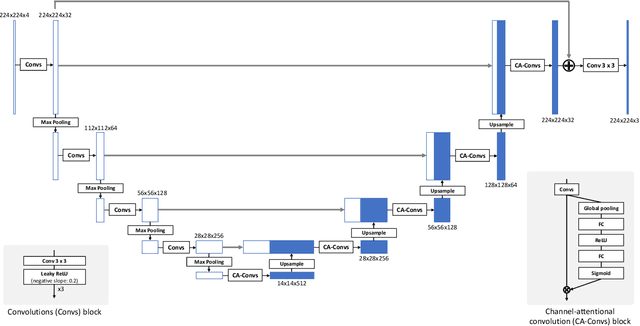
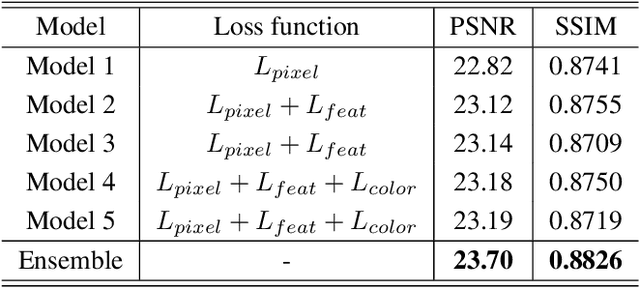
Abstract:Recent research on learning a mapping between raw Bayer images and RGB images has progressed with the development of deep convolutional neural networks. A challenging data set namely the Zurich Raw-to-RGB data set (ZRR) has been released in the AIM 2019 raw-to-RGB mapping challenge. In ZRR, input raw and target RGB images are captured by two different cameras and thus not perfectly aligned. Moreover, camera metadata such as white balance gains and color correction matrix are not provided, which makes the challenge more difficult. In this paper, we explore an effective network structure and a loss function to address these issues. We exploit a two-stage U-Net architecture and also introduce a loss function that is less variant to alignment and more sensitive to color differences. In addition, we show an ensemble of networks trained with different loss functions can bring a significant performance gain. We demonstrate the superiority of our method by achieving the highest score in terms of both the peak signal-to-noise ratio and the structural similarity and obtaining the second-best mean-opinion-score in the challenge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge