Kapil Ahuja
Chameleon2++: An Efficient Chameleon2 Clustering with Approximate Nearest Neighbors
Jan 05, 2025Abstract:Clustering algorithms are fundamental tools in data analysis, with hierarchical methods being particularly valuable for their flexibility. Chameleon is a widely used hierarchical clustering algorithm that excels at identifying high-quality clusters of arbitrary shapes, sizes, and densities. Chameleon2 is the most recent variant that has demonstrated significant improvements, but suffers from critical failings and there are certain improvements that can be made. The first failure we address is that the complexity of Chameleon2 is claimed to be $O(n^2)$, while we demonstrate that it is actually $O(n^2\log{n})$, with $n$ being the number of data points. Furthermore, we suggest improvements to Chameleon2 that ensure that the complexity remains $O(n^2)$ with minimal to no loss of performance. The second failing of Chameleon2 is that it lacks transparency and it does not provide the fine-tuned algorithm parameters used to obtain the claimed results. We meticulously provide all such parameter values to enhance replicability. The improvement which we make in Chameleon2 is that we replace the exact $k$-NN search with an approximate $k$-NN search. This further reduces the algorithmic complexity down to $O(n\log{n})$ without any performance loss. Here, we primarily configure three approximate nearest neighbor search algorithms (Annoy, FLANN and NMSLIB) to align with the overarching Chameleon2 clustering framework. Experimental evaluations on standard benchmark datasets demonstrate that the proposed Chameleon2++ algorithm is more efficient, robust, and computationally optimal.
AI Algorithm for Predicting and Optimizing Trajectory of UAV Swarm
May 20, 2024Abstract:This paper explores the application of Artificial Intelligence (AI) techniques for generating the trajectories of fleets of Unmanned Aerial Vehicles (UAVs). The two main challenges addressed include accurately predicting the paths of UAVs and efficiently avoiding collisions between them. Firstly, the paper systematically applies a diverse set of activation functions to a Feedforward Neural Network (FFNN) with a single hidden layer, which enhances the accuracy of the predicted path compared to previous work. Secondly, we introduce a novel activation function, AdaptoSwelliGauss, which is a sophisticated fusion of Swish and Elliott activations, seamlessly integrated with a scaled and shifted Gaussian component. Swish facilitates smooth transitions, Elliott captures abrupt trajectory changes, and the scaled and shifted Gaussian enhances robustness against noise. This dynamic combination is specifically designed to excel in capturing the complexities of UAV trajectory prediction. This new activation function gives substantially better accuracy than all existing activation functions. Thirdly, we propose a novel Integrated Collision Detection, Avoidance, and Batching (ICDAB) strategy that merges two complementary UAV collision avoidance techniques: changing UAV trajectories and altering their starting times, also referred to as batching. This integration helps overcome the disadvantages of both - reduction in the number of trajectory manipulations, which avoids overly convoluted paths in the first technique, and smaller batch sizes, which reduce overall takeoff time in the second.
Deep Learning Descriptor Hybridization with Feature Reduction for Accurate Cervical Cancer Colposcopy Image Classification
May 01, 2024Abstract:Cervical cancer stands as a predominant cause of female mortality, underscoring the need for regular screenings to enable early diagnosis and preemptive treatment of pre-cancerous conditions. The transformation zone in the cervix, where cellular differentiation occurs, plays a critical role in the detection of abnormalities. Colposcopy has emerged as a pivotal tool in cervical cancer prevention since it provides a meticulous examination of cervical abnormalities. However, challenges in visual evaluation necessitate the development of Computer Aided Diagnosis (CAD) systems. We propose a novel CAD system that combines the strengths of various deep-learning descriptors (ResNet50, ResNet101, and ResNet152) with appropriate feature normalization (min-max) as well as feature reduction technique (LDA). The combination of different descriptors ensures that all the features (low-level like edges and colour, high-level like shape and texture) are captured, feature normalization prevents biased learning, and feature reduction avoids overfitting. We do experiments on the IARC dataset provided by WHO. The dataset is initially segmented and balanced. Our approach achieves exceptional performance in the range of 97%-100% for both the normal-abnormal and the type classification. A competitive approach for type classification on the same dataset achieved 81%-91% performance.
A Novel Sampled Clustering Algorithm for Rice Phenotypic Data
Dec 22, 2023Abstract:Phenotypic (or Physical) characteristics of plant species are commonly used to perform clustering. In one of our recent works (Shastri et al. (2021)), we used a probabilistically sampled (using pivotal sampling) and spectrally clustered algorithm to group soybean species. These techniques were used to obtain highly accurate clusterings at a reduced cost. In this work, we extend the earlier algorithm to cluster rice species. We improve the base algorithm in three ways. First, we propose a new function to build the similarity matrix in Spectral Clustering. Commonly, a natural exponential function is used for this purpose. Based upon the spectral graph theory and the involved Cheeger's inequality, we propose the use a base "a" exponential function instead. This gives a similarity matrix spectrum favorable for clustering, which we support via an eigenvalue analysis. Second, the function used to build the similarity matrix in Spectral Clustering was earlier scaled with a fixed factor (called global scaling). Based upon the idea of Zelnik-Manor and Perona (2004), we now use a factor that varies with matrix elements (called local scaling) and works better. Third, to compute the inclusion probability of a specie in the pivotal sampling algorithm, we had earlier used the notion of deviation that captured how far specie's characteristic values were from their respective base values (computed over all species). A maximum function was used before to find the base values. We now use a median function, which is more intuitive. We support this choice using a statistical analysis. With experiments on 1865 rice species, we demonstrate that in terms of silhouette values, our new Sampled Spectral Clustering is 61% better than Hierarchical Clustering (currently prevalent). Also, our new algorithm is significantly faster than Hierarchical Clustering due to the involved sampling.
Cube Sampled K-Prototype Clustering for Featured Data
Aug 23, 2021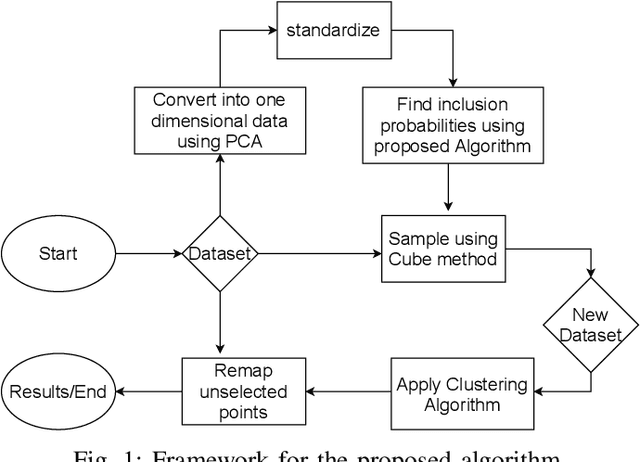
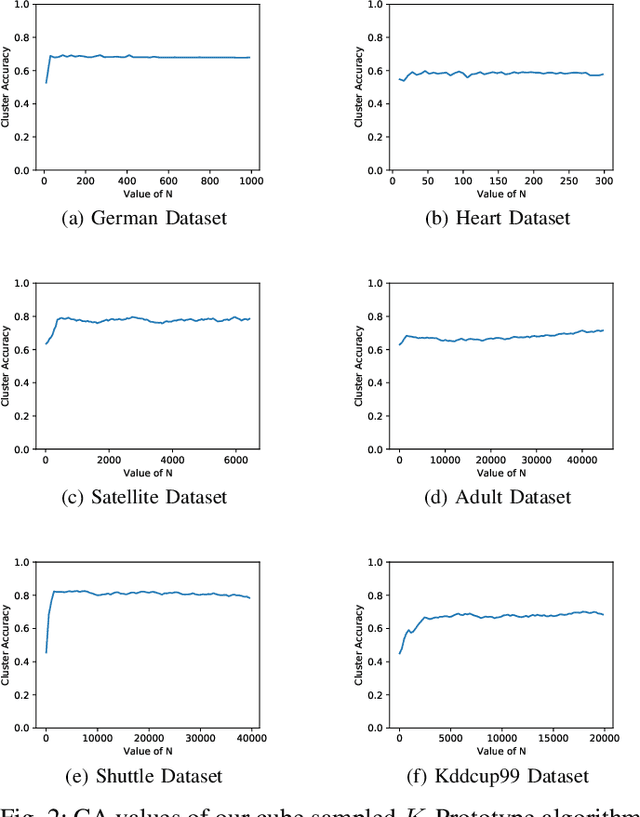
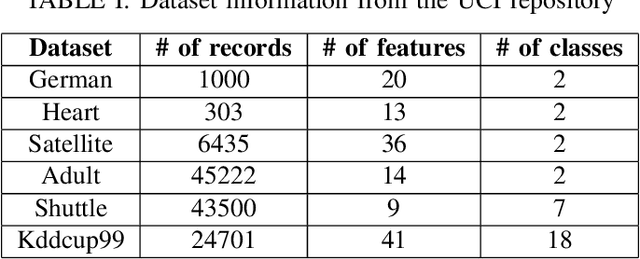
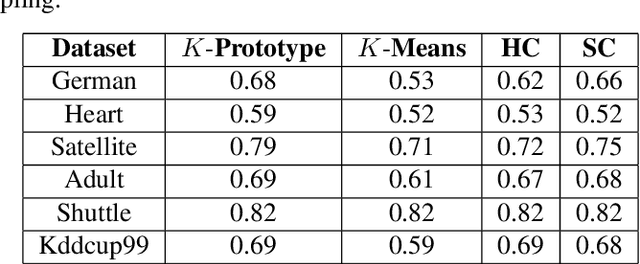
Abstract:Clustering large amount of data is becoming increasingly important in the current times. Due to the large sizes of data, clustering algorithm often take too much time. Sampling this data before clustering is commonly used to reduce this time. In this work, we propose a probabilistic sampling technique called cube sampling along with K-Prototype clustering. Cube sampling is used because of its accurate sample selection. K-Prototype is most frequently used clustering algorithm when the data is numerical as well as categorical (very common in today's time). The novelty of this work is in obtaining the crucial inclusion probabilities for cube sampling using Principal Component Analysis (PCA). Experiments on multiple datasets from the UCI repository demonstrate that cube sampled K-Prototype algorithm gives the best clustering accuracy among similarly sampled other popular clustering algorithms (K-Means, Hierarchical Clustering (HC), Spectral Clustering (SC)). When compared with unsampled K-Prototype, K-Means, HC and SC, it still has the best accuracy with the added advantage of reduced computational complexity (due to reduced data size).
Probabilistically Sampled and Spectrally Clustered Plant Genotypes using Phenotypic Characteristics
Sep 18, 2020
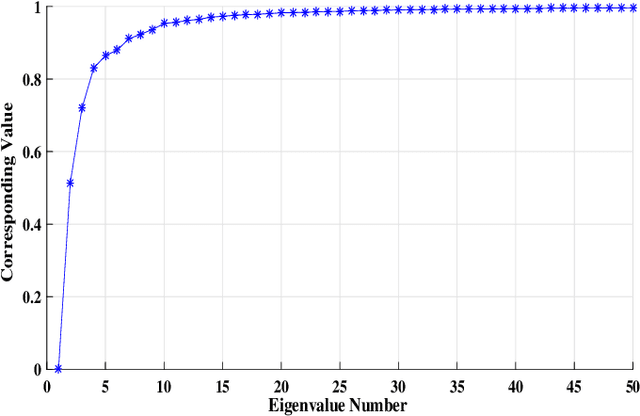
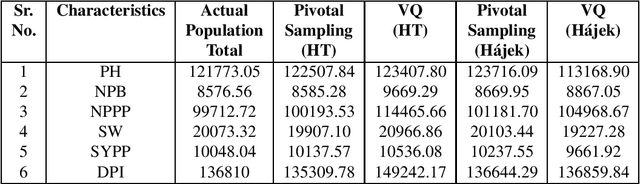
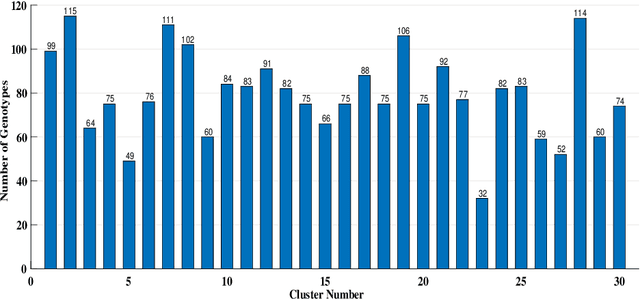
Abstract:Clustering genotypes based upon their phenotypic characteristics is used to obtain diverse sets of parents that are useful in their breeding programs. The Hierarchical Clustering (HC) algorithm is the current standard in clustering of phenotypic data. This algorithm suffers from low accuracy and high computational complexity issues. To address the accuracy challenge, we propose the use of Spectral Clustering (SC) algorithm. To make the algorithm computationally cheap, we propose using sampling, specifically, Pivotal Sampling that is probability based. Since application of samplings to phenotypic data has not been explored much, for effective comparison, another sampling technique called Vector Quantization (VQ) is adapted for this data as well. VQ has recently given promising results for genome data. The novelty of our SC with Pivotal Sampling algorithm is in constructing the crucial similarity matrix for the clustering algorithm and defining probabilities for the sampling technique. Although our algorithm can be applied to any plant genotypes, we test it on the phenotypic data obtained from about 2400 Soybean genotypes. SC with Pivotal Sampling achieves substantially more accuracy (in terms of Silhouette Values) than all the other proposed competitive clustering with sampling algorithms (i.e. SC with VQ, HC with Pivotal Sampling, and HC with VQ). The complexities of our SC with Pivotal Sampling algorithm and these three variants are almost same because of the involved sampling. In addition to this, SC with Pivotal Sampling outperforms the standard HC algorithm in both accuracy and computational complexity. We experimentally show that we are up to 45% more accurate than HC in terms of clustering accuracy. The computational complexity of our algorithm is more than a magnitude lesser than HC.
Vector Quantized Spectral Clustering applied to Soybean Whole Genome Sequences
Sep 30, 2018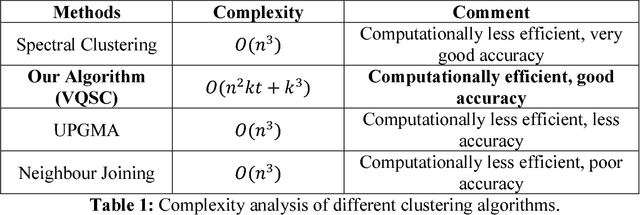
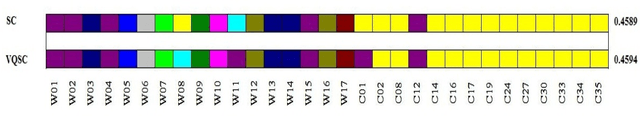
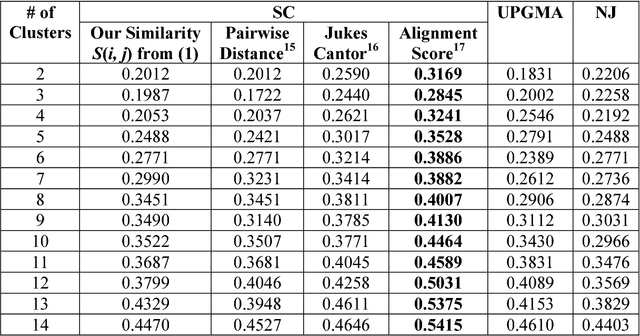
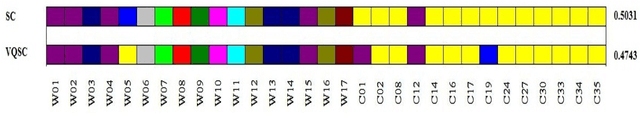
Abstract:We develop a Vector Quantized Spectral Clustering (VQSC) algorithm that is a combination of Spectral Clustering (SC) and Vector Quantization (VQ) sampling for grouping Soybean genomes. The inspiration here is to use SC for its accuracy and VQ to make the algorithm computationally cheap (the complexity of SC is cubic in-terms of the input size). Although the combination of SC and VQ is not new, the novelty of our work is in developing the crucial similarity matrix in SC as well as use of k-medoids in VQ, both adapted for the Soybean genome data. We compare our approach with commonly used techniques like UPGMA (Un-weighted Pair Graph Method with Arithmetic Mean) and NJ (Neighbour Joining). Experimental results show that our approach outperforms both these techniques significantly in terms of cluster quality (up to 25% better cluster quality) and time complexity (order of magnitude faster).
Localized Multiple Kernel Learning for Anomaly Detection: One-class Classification
Jul 17, 2018



Abstract:Multi-kernel learning has been well explored in the recent past and has exhibited promising outcomes for multi-class classification and regression tasks. In this paper, we present a multiple kernel learning approach for the One-class Classification (OCC) task and employ it for anomaly detection. Recently, the basic multi-kernel approach has been proposed to solve the OCC problem, which is simply a convex combination of different kernels with equal weights. This paper proposes a Localized Multiple Kernel learning approach for Anomaly Detection (LMKAD) using OCC, where the weight for each kernel is assigned locally. Proposed LMKAD approach adapts the weight for each kernel using a gating function. The parameters of the gating function and one-class classifier are optimized simultaneously through a two-step optimization process. We present the empirical results of the performance of LMKAD on 25 benchmark datasets from various disciplines. This performance is evaluated against existing Multi Kernel Anomaly Detection (MKAD) algorithm, and four other existing kernel-based one-class classifiers to showcase the credibility of our approach. Our algorithm achieves significantly better Gmean scores while using a lesser number of support vectors compared to MKAD. Friedman test is also performed to verify the statistical significance of the results claimed in this paper.
Online Learning with Regularized Kernel for One-class Classification
Apr 09, 2018



Abstract:This paper presents an online learning with regularized kernel based one-class extreme learning machine (ELM) classifier and is referred as online RK-OC-ELM. The baseline kernel hyperplane model considers whole data in a single chunk with regularized ELM approach for offline learning in case of one-class classification (OCC). Further, the basic hyper plane model is adapted in an online fashion from stream of training samples in this paper. Two frameworks viz., boundary and reconstruction are presented to detect the target class in online RKOC-ELM. Boundary framework based one-class classifier consists of single node output architecture and classifier endeavors to approximate all data to any real number. However, one-class classifier based on reconstruction framework is an autoencoder architecture, where output nodes are identical to input nodes and classifier endeavor to reconstruct input layer at the output layer. Both these frameworks employ regularized kernel ELM based online learning and consistency based model selection has been employed to select learning algorithm parameters. The performance of online RK-OC-ELM has been evaluated on standard benchmark datasets as well as on artificial datasets and the results are compared with existing state-of-the art one-class classifiers. The results indicate that the online learning one-class classifier is slightly better or same as batch learning based approaches. As, base classifier used for the proposed classifiers are based on the ELM, hence, proposed classifiers would also inherit the benefit of the base classifier i.e. it will perform faster computation compared to traditional autoencoder based one-class classifier.
Density-Wise Two Stage Mammogram Classification using Texture Exploiting Descriptors
Jan 03, 2018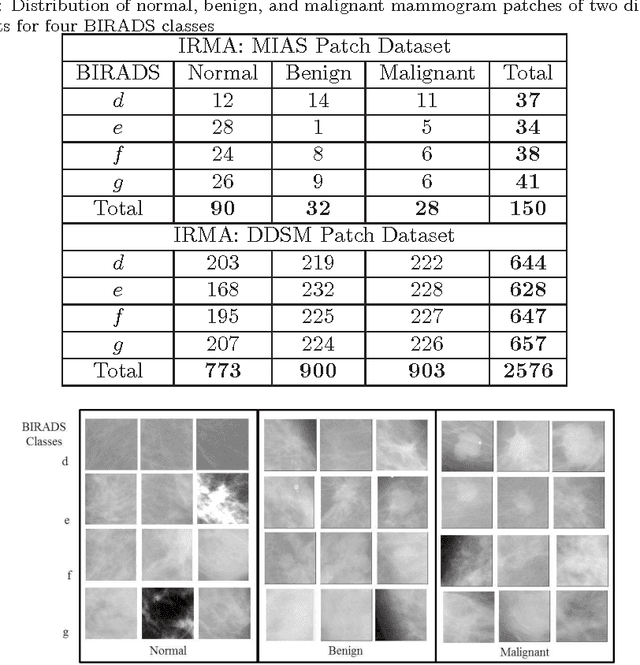
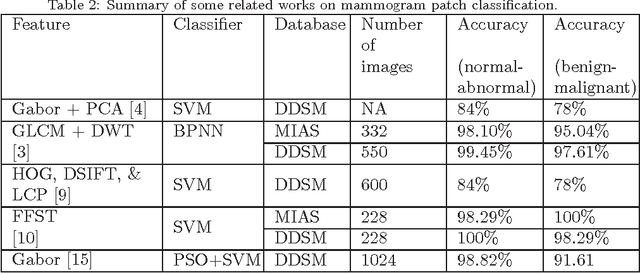
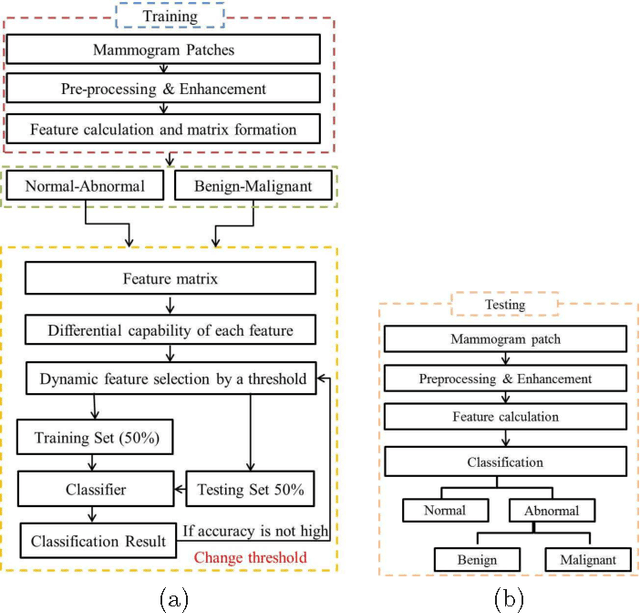
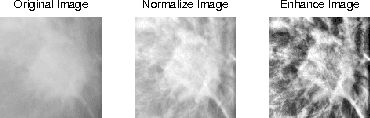
Abstract:Breast cancer is becoming pervasive with each passing day. Hence, its early detection is a big step in saving the life of any patient. Mammography is a common tool in breast cancer diagnosis. The most important step here is classification of mammogram patches as normal-abnormal and benign-malignant. Texture of a breast in a mammogram patch plays a significant role in these classifications. We propose a variation of Histogram of Gradients (HOG) and Gabor filter combination called Histogram of Oriented Texture (HOT) that exploits this fact. We also revisit the Pass Band - Discrete Cosine Transform (PB-DCT) descriptor that captures texture information well. All features of a mammogram patch may not be useful. Hence, we apply a feature selection technique called Discrimination Potentiality (DP). Our resulting descriptors, DP-HOT and DP-PB-DCT, are compared with the standard descriptors. Density of a mammogram patch is important for classification, and has not been studied exhaustively. The Image Retrieval in Medical Application (IRMA) database from RWTH Aachen, Germany is a standard database that provides mammogram patches, and most researchers have tested their frameworks only on a subset of patches from this database. We apply our two new descriptors on all images of the IRMA database for density wise classification, and compare with the standard descriptors. We achieve higher accuracy than all of the existing standard descriptors (more than 92%).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge