Aditya A. Shastri
Cube Sampled K-Prototype Clustering for Featured Data
Aug 23, 2021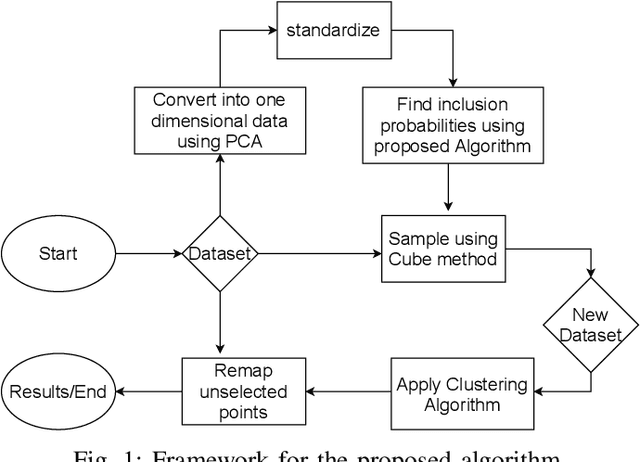
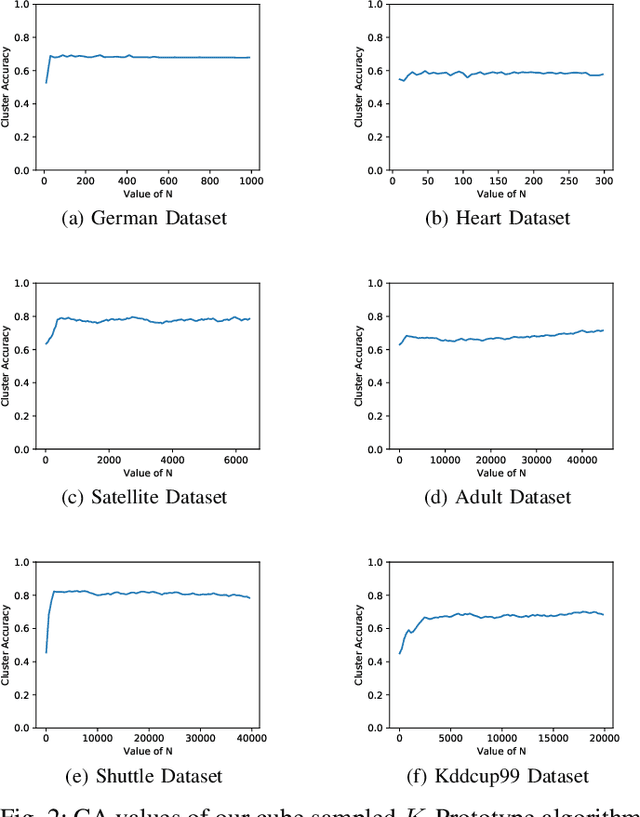
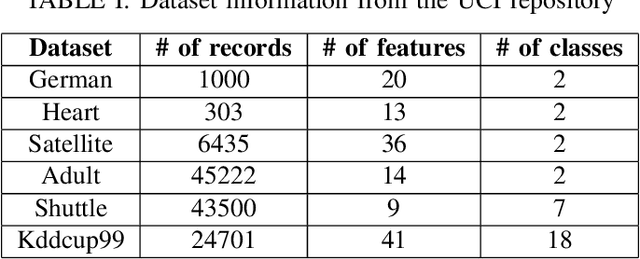
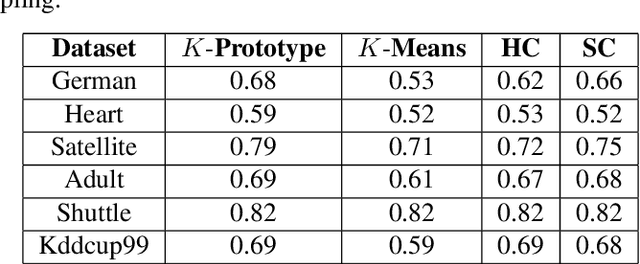
Abstract:Clustering large amount of data is becoming increasingly important in the current times. Due to the large sizes of data, clustering algorithm often take too much time. Sampling this data before clustering is commonly used to reduce this time. In this work, we propose a probabilistic sampling technique called cube sampling along with K-Prototype clustering. Cube sampling is used because of its accurate sample selection. K-Prototype is most frequently used clustering algorithm when the data is numerical as well as categorical (very common in today's time). The novelty of this work is in obtaining the crucial inclusion probabilities for cube sampling using Principal Component Analysis (PCA). Experiments on multiple datasets from the UCI repository demonstrate that cube sampled K-Prototype algorithm gives the best clustering accuracy among similarly sampled other popular clustering algorithms (K-Means, Hierarchical Clustering (HC), Spectral Clustering (SC)). When compared with unsampled K-Prototype, K-Means, HC and SC, it still has the best accuracy with the added advantage of reduced computational complexity (due to reduced data size).
Probabilistically Sampled and Spectrally Clustered Plant Genotypes using Phenotypic Characteristics
Sep 18, 2020
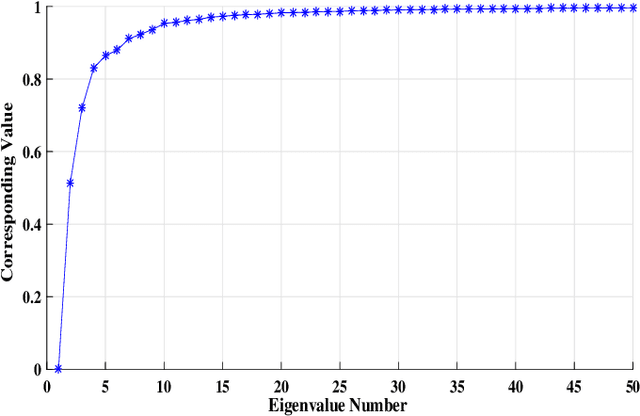
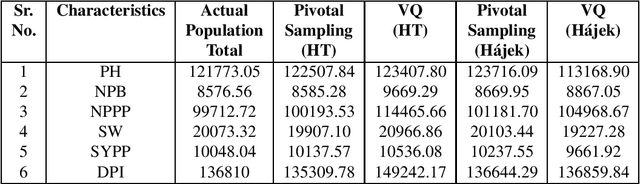
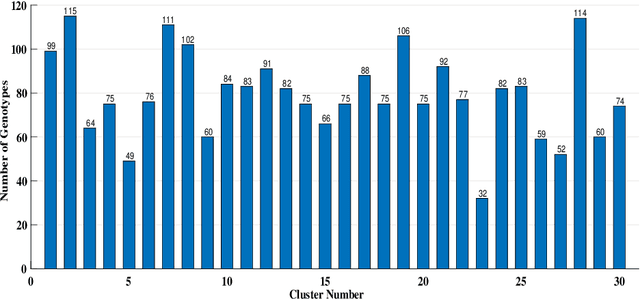
Abstract:Clustering genotypes based upon their phenotypic characteristics is used to obtain diverse sets of parents that are useful in their breeding programs. The Hierarchical Clustering (HC) algorithm is the current standard in clustering of phenotypic data. This algorithm suffers from low accuracy and high computational complexity issues. To address the accuracy challenge, we propose the use of Spectral Clustering (SC) algorithm. To make the algorithm computationally cheap, we propose using sampling, specifically, Pivotal Sampling that is probability based. Since application of samplings to phenotypic data has not been explored much, for effective comparison, another sampling technique called Vector Quantization (VQ) is adapted for this data as well. VQ has recently given promising results for genome data. The novelty of our SC with Pivotal Sampling algorithm is in constructing the crucial similarity matrix for the clustering algorithm and defining probabilities for the sampling technique. Although our algorithm can be applied to any plant genotypes, we test it on the phenotypic data obtained from about 2400 Soybean genotypes. SC with Pivotal Sampling achieves substantially more accuracy (in terms of Silhouette Values) than all the other proposed competitive clustering with sampling algorithms (i.e. SC with VQ, HC with Pivotal Sampling, and HC with VQ). The complexities of our SC with Pivotal Sampling algorithm and these three variants are almost same because of the involved sampling. In addition to this, SC with Pivotal Sampling outperforms the standard HC algorithm in both accuracy and computational complexity. We experimentally show that we are up to 45% more accurate than HC in terms of clustering accuracy. The computational complexity of our algorithm is more than a magnitude lesser than HC.
Vector Quantized Spectral Clustering applied to Soybean Whole Genome Sequences
Sep 30, 2018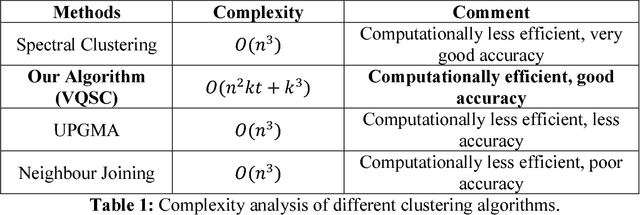
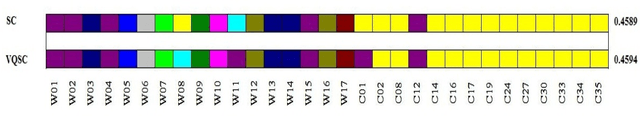
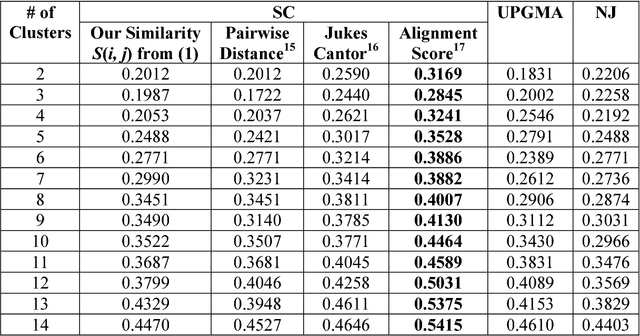
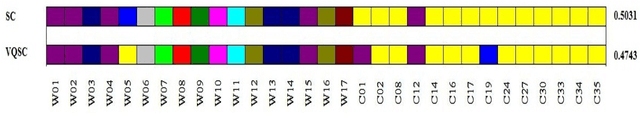
Abstract:We develop a Vector Quantized Spectral Clustering (VQSC) algorithm that is a combination of Spectral Clustering (SC) and Vector Quantization (VQ) sampling for grouping Soybean genomes. The inspiration here is to use SC for its accuracy and VQ to make the algorithm computationally cheap (the complexity of SC is cubic in-terms of the input size). Although the combination of SC and VQ is not new, the novelty of our work is in developing the crucial similarity matrix in SC as well as use of k-medoids in VQ, both adapted for the Soybean genome data. We compare our approach with commonly used techniques like UPGMA (Un-weighted Pair Graph Method with Arithmetic Mean) and NJ (Neighbour Joining). Experimental results show that our approach outperforms both these techniques significantly in terms of cluster quality (up to 25% better cluster quality) and time complexity (order of magnitude faster).
Density-Wise Two Stage Mammogram Classification using Texture Exploiting Descriptors
Jan 03, 2018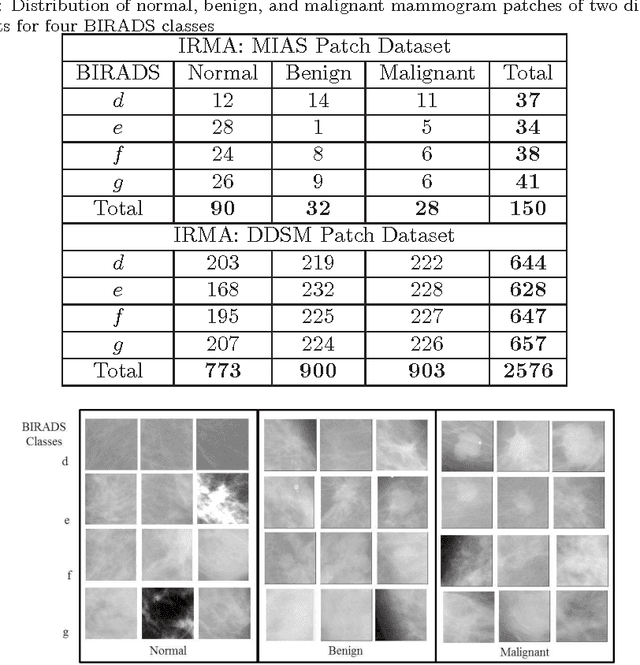
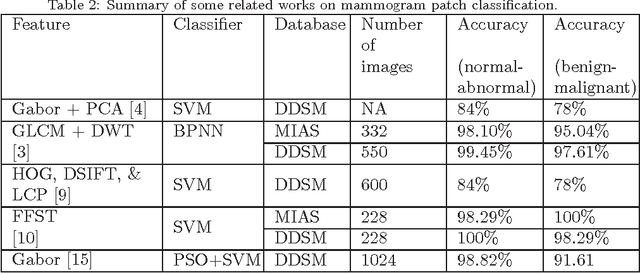
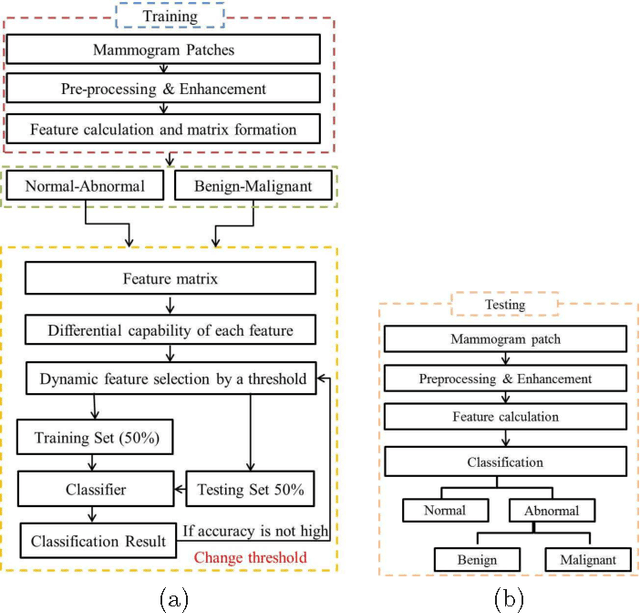
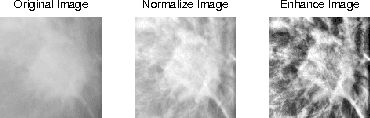
Abstract:Breast cancer is becoming pervasive with each passing day. Hence, its early detection is a big step in saving the life of any patient. Mammography is a common tool in breast cancer diagnosis. The most important step here is classification of mammogram patches as normal-abnormal and benign-malignant. Texture of a breast in a mammogram patch plays a significant role in these classifications. We propose a variation of Histogram of Gradients (HOG) and Gabor filter combination called Histogram of Oriented Texture (HOT) that exploits this fact. We also revisit the Pass Band - Discrete Cosine Transform (PB-DCT) descriptor that captures texture information well. All features of a mammogram patch may not be useful. Hence, we apply a feature selection technique called Discrimination Potentiality (DP). Our resulting descriptors, DP-HOT and DP-PB-DCT, are compared with the standard descriptors. Density of a mammogram patch is important for classification, and has not been studied exhaustively. The Image Retrieval in Medical Application (IRMA) database from RWTH Aachen, Germany is a standard database that provides mammogram patches, and most researchers have tested their frameworks only on a subset of patches from this database. We apply our two new descriptors on all images of the IRMA database for density wise classification, and compare with the standard descriptors. We achieve higher accuracy than all of the existing standard descriptors (more than 92%).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge