Juntao Jiang
College of Control Science and Engineering, Zhejiang University, Hangzhou, China
FUGC: Benchmarking Semi-Supervised Learning Methods for Cervical Segmentation
Jan 22, 2026Abstract:Accurate segmentation of cervical structures in transvaginal ultrasound (TVS) is critical for assessing the risk of spontaneous preterm birth (PTB), yet the scarcity of labeled data limits the performance of supervised learning approaches. This paper introduces the Fetal Ultrasound Grand Challenge (FUGC), the first benchmark for semi-supervised learning in cervical segmentation, hosted at ISBI 2025. FUGC provides a dataset of 890 TVS images, including 500 training images, 90 validation images, and 300 test images. Methods were evaluated using the Dice Similarity Coefficient (DSC), Hausdorff Distance (HD), and runtime (RT), with a weighted combination of 0.4/0.4/0.2. The challenge attracted 10 teams with 82 participants submitting innovative solutions. The best-performing methods for each individual metric achieved 90.26\% mDSC, 38.88 mHD, and 32.85 ms RT, respectively. FUGC establishes a standardized benchmark for cervical segmentation, demonstrates the efficacy of semi-supervised methods with limited labeled data, and provides a foundation for AI-assisted clinical PTB risk assessment.
Large-Scale Multidimensional Knowledge Profiling of Scientific Literature
Jan 21, 2026Abstract:The rapid expansion of research across machine learning, vision, and language has produced a volume of publications that is increasingly difficult to synthesize. Traditional bibliometric tools rely mainly on metadata and offer limited visibility into the semantic content of papers, making it hard to track how research themes evolve over time or how different areas influence one another. To obtain a clearer picture of recent developments, we compile a unified corpus of more than 100,000 papers from 22 major conferences between 2020 and 2025 and construct a multidimensional profiling pipeline to organize and analyze their textual content. By combining topic clustering, LLM-assisted parsing, and structured retrieval, we derive a comprehensive representation of research activity that supports the study of topic lifecycles, methodological transitions, dataset and model usage patterns, and institutional research directions. Our analysis highlights several notable shifts, including the growth of safety, multimodal reasoning, and agent-oriented studies, as well as the gradual stabilization of areas such as neural machine translation and graph-based methods. These findings provide an evidence-based view of how AI research is evolving and offer a resource for understanding broader trends and identifying emerging directions. Code and dataset: https://github.com/xzc-zju/Profiling_Scientific_Literature
M3CoTBench: Benchmark Chain-of-Thought of MLLMs in Medical Image Understanding
Jan 13, 2026Abstract:Chain-of-Thought (CoT) reasoning has proven effective in enhancing large language models by encouraging step-by-step intermediate reasoning, and recent advances have extended this paradigm to Multimodal Large Language Models (MLLMs). In the medical domain, where diagnostic decisions depend on nuanced visual cues and sequential reasoning, CoT aligns naturally with clinical thinking processes. However, Current benchmarks for medical image understanding generally focus on the final answer while ignoring the reasoning path. An opaque process lacks reliable bases for judgment, making it difficult to assist doctors in diagnosis. To address this gap, we introduce a new M3CoTBench benchmark specifically designed to evaluate the correctness, efficiency, impact, and consistency of CoT reasoning in medical image understanding. M3CoTBench features 1) a diverse, multi-level difficulty dataset covering 24 examination types, 2) 13 varying-difficulty tasks, 3) a suite of CoT-specific evaluation metrics (correctness, efficiency, impact, and consistency) tailored to clinical reasoning, and 4) a performance analysis of multiple MLLMs. M3CoTBench systematically evaluates CoT reasoning across diverse medical imaging tasks, revealing current limitations of MLLMs in generating reliable and clinically interpretable reasoning, and aims to foster the development of transparent, trustworthy, and diagnostically accurate AI systems for healthcare. Project page at https://juntaojianggavin.github.io/projects/M3CoTBench/.
UltraLBM-UNet: Ultralight Bidirectional Mamba-based Model for Skin Lesion Segmentation
Dec 25, 2025Abstract:Skin lesion segmentation is a crucial step in dermatology for guiding clinical decision-making. However, existing methods for accurate, robust, and resource-efficient lesion analysis have limitations, including low performance and high computational complexity. To address these limitations, we propose UltraLBM-UNet, a lightweight U-Net variant that integrates a bidirectional Mamba-based global modeling mechanism with multi-branch local feature perception. The proposed architecture integrates efficient local feature injection with bidirectional state-space modeling, enabling richer contextual interaction across spatial dimensions while maintaining computational compactness suitable for point-of-care deployment. Extensive experiments on the ISIC 2017, ISIC 2018, and PH2 datasets demonstrate that our model consistently achieves state-of-the-art segmentation accuracy, outperforming existing lightweight and Mamba counterparts with only 0.034M parameters and 0.060 GFLOPs. In addition, we introduce a hybrid knowledge distillation strategy to train an ultra-compact student model, where the distilled variant UltraLBM-UNet-T, with only 0.011M parameters and 0.019 GFLOPs, achieves competitive segmentation performance. These results highlight the suitability of UltraLBM-UNet for point-of-care deployment, where accurate and robust lesion analyses are essential. The source code is publicly available at https://github.com/LinLinLin-X/UltraLBM-UNet.
VisAlgae 2023: A Dataset and Challenge for Algae Detection in Microscopy Images
May 27, 2025Abstract:Microalgae, vital for ecological balance and economic sectors, present challenges in detection due to their diverse sizes and conditions. This paper summarizes the second "Vision Meets Algae" (VisAlgae 2023) Challenge, aiming to enhance high-throughput microalgae cell detection. The challenge, which attracted 369 participating teams, includes a dataset of 1000 images across six classes, featuring microalgae of varying sizes and distinct features. Participants faced tasks such as detecting small targets, handling motion blur, and complex backgrounds. The top 10 methods, outlined here, offer insights into overcoming these challenges and maximizing detection accuracy. This intersection of algae research and computer vision offers promise for ecological understanding and technological advancement. The dataset can be accessed at: https://github.com/juntaoJianggavin/Visalgae2023/.
DiffuMural: Restoring Dunhuang Murals with Multi-scale Diffusion
Apr 13, 2025Abstract:Large-scale pre-trained diffusion models have produced excellent results in the field of conditional image generation. However, restoration of ancient murals, as an important downstream task in this field, poses significant challenges to diffusion model-based restoration methods due to its large defective area and scarce training samples. Conditional restoration tasks are more concerned with whether the restored part meets the aesthetic standards of mural restoration in terms of overall style and seam detail, and such metrics for evaluating heuristic image complements are lacking in current research. We therefore propose DiffuMural, a combined Multi-scale convergence and Collaborative Diffusion mechanism with ControlNet and cyclic consistency loss to optimise the matching between the generated images and the conditional control. DiffuMural demonstrates outstanding capabilities in mural restoration, leveraging training data from 23 large-scale Dunhuang murals that exhibit consistent visual aesthetics. The model excels in restoring intricate details, achieving a coherent overall appearance, and addressing the unique challenges posed by incomplete murals lacking factual grounding. Our evaluation framework incorporates four key metrics to quantitatively assess incomplete murals: factual accuracy, textural detail, contextual semantics, and holistic visual coherence. Furthermore, we integrate humanistic value assessments to ensure the restored murals retain their cultural and artistic significance. Extensive experiments validate that our method outperforms state-of-the-art (SOTA) approaches in both qualitative and quantitative metrics.
RWKV-UNet: Improving UNet with Long-Range Cooperation for Effective Medical Image Segmentation
Jan 14, 2025



Abstract:In recent years, there have been significant advancements in deep learning for medical image analysis, especially with convolutional neural networks (CNNs) and transformer models. However, CNNs face limitations in capturing long-range dependencies while transformers suffer high computational complexities. To address this, we propose RWKV-UNet, a novel model that integrates the RWKV (Receptance Weighted Key Value) structure into the U-Net architecture. This integration enhances the model's ability to capture long-range dependencies and improve contextual understanding, which is crucial for accurate medical image segmentation. We build a strong encoder with developed inverted residual RWKV (IR-RWKV) blocks combining CNNs and RWKVs. We also propose a Cross-Channel Mix (CCM) module to improve skip connections with multi-scale feature fusion, achieving global channel information integration. Experiments on benchmark datasets, including Synapse, ACDC, BUSI, CVC-ClinicDB, CVC-ColonDB, Kvasir-SEG, ISIC 2017 and GLAS show that RWKV-UNet achieves state-of-the-art performance on various types of medical image segmentation. Additionally, smaller variants, RWKV-UNet-S and RWKV-UNet-T, balance accuracy and computational efficiency, making them suitable for broader clinical applications.
LV-UNet: A Lightweight and Vanilla Model for Medical Image Segmentation
Aug 29, 2024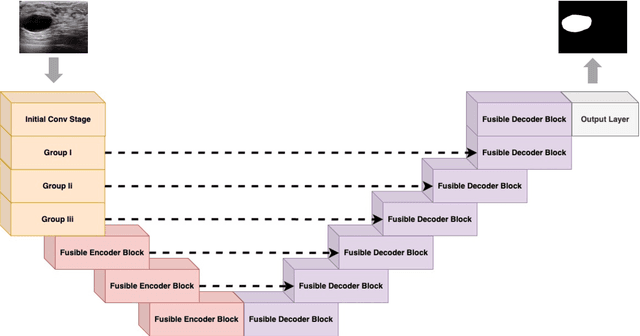
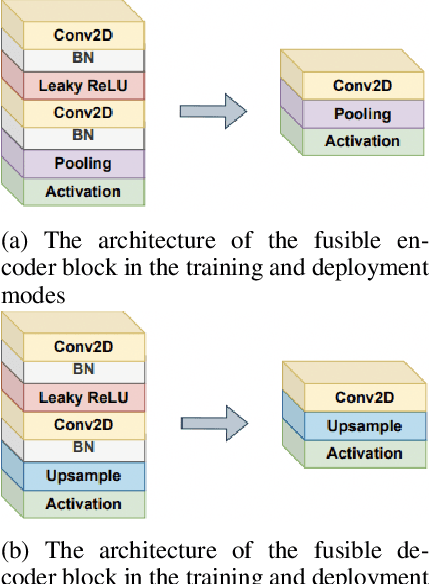
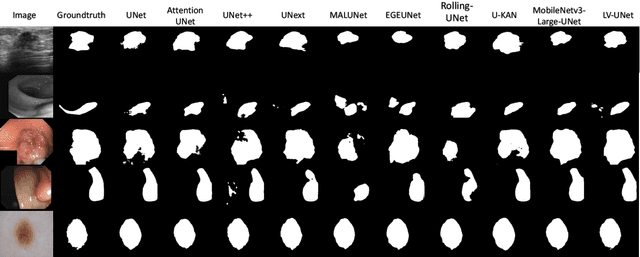
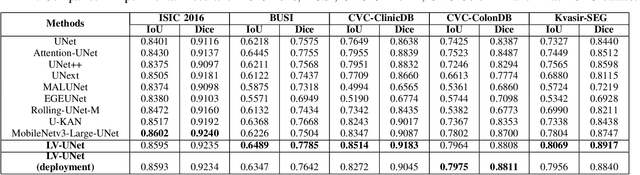
Abstract:Although the progress made by large models in computer vision, optimization challenges, the complexity of transformer models, computational limitations, and the requirements of practical applications call for simpler designs in model architecture for medical image segmentation, especially in mobile medical devices that require lightweight and deployable models with real-time performance. However, some of the current lightweight models exhibit poor robustness across different datasets, which hinders their broader adoption. This paper proposes a lightweight and vanilla model called LV-UNet, which effectively utilizes pre-trained MobileNetv3-Large models and introduces fusible modules. It can be trained using an improved deep training strategy and switched to deployment mode during inference, reducing both parameter count and computational load. Experiments are conducted on ISIC 2016, BUSI, CVC- ClinicDB, CVC-ColonDB, and Kvair-SEG datasets, achieving better performance compared to the state-of-the-art and classic models.
ViG-UNet: Vision Graph Neural Networks for Medical Image Segmentation
Jun 08, 2023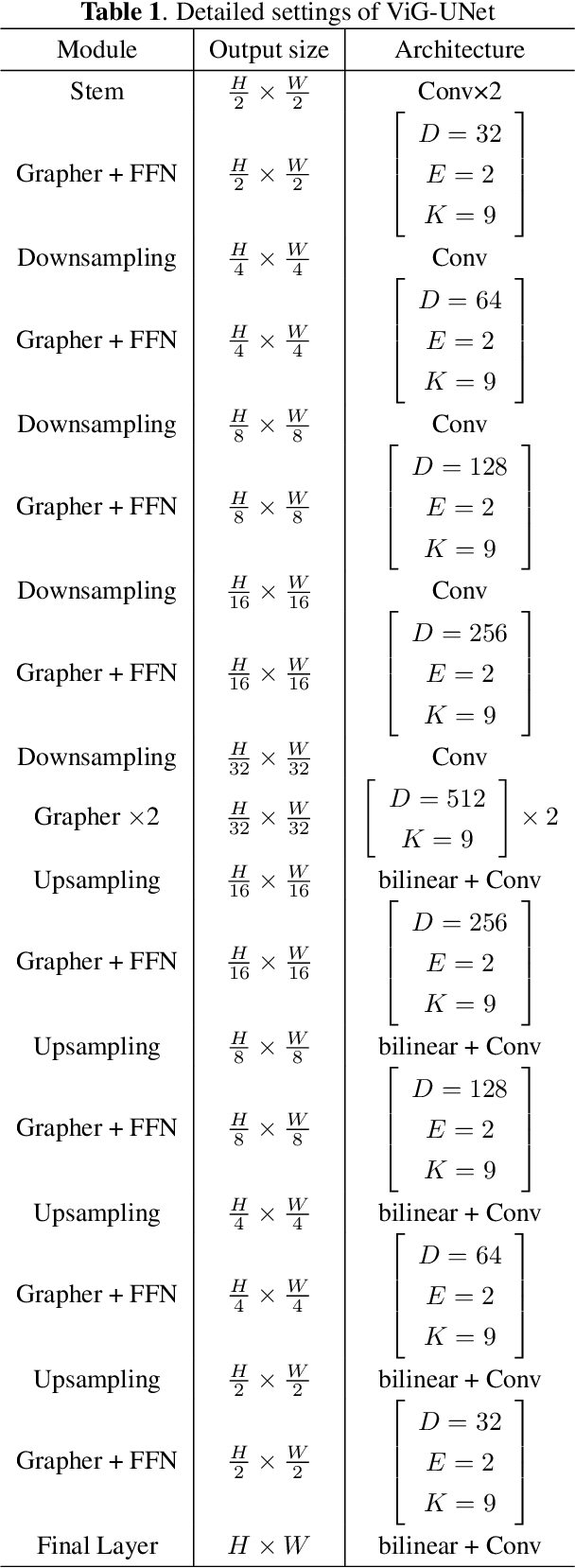
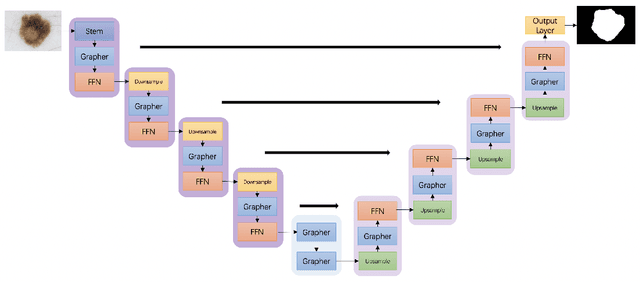
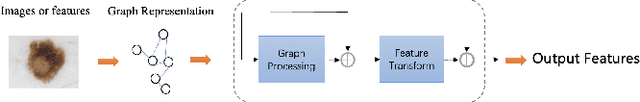
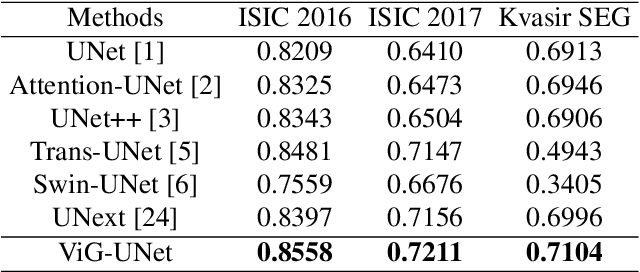
Abstract:Deep neural networks have been widely used in medical image analysis and medical image segmentation is one of the most important tasks. U-shaped neural networks with encoder-decoder are prevailing and have succeeded greatly in various segmentation tasks. While CNNs treat an image as a grid of pixels in Euclidean space and Transformers recognize an image as a sequence of patches, graph-based representation is more generalized and can construct connections for each part of an image. In this paper, we propose a novel ViG-UNet, a graph neural network-based U-shaped architecture with the encoder, the decoder, the bottleneck, and skip connections. The downsampling and upsampling modules are also carefully designed. The experimental results on ISIC 2016, ISIC 2017 and Kvasir-SEG datasets demonstrate that our proposed architecture outperforms most existing classic and state-of-the-art U-shaped networks.
Marine Microalgae Detection in Microscopy Images: A New Dataset
Nov 14, 2022



Abstract:Marine microalgae are widespread in the ocean and play a crucial role in the ecosystem. Automatic identification and location of marine microalgae in microscopy images would help establish marine ecological environment monitoring and water quality evaluation system. A new dataset for marine microalgae detection is proposed in this paper. Six classes of microalgae commonlyfound in the ocean (Bacillariophyta, Chlorella pyrenoidosa, Platymonas, Dunaliella salina, Chrysophyta, Symbiodiniaceae) are microscopically imaged in real-time. Images of Symbiodiniaceae in three physiological states known as normal, bleaching, and translating are also included. We annotated these images with bounding boxes using Labelme software and split them into the training and testing sets. The total number of images in the dataset is 937 and all the objects in these images were annotated. The total number of annotated objects is 4201. The training set contains 537 images and the testing set contains 430 images. Baselines of different object detection algorithms are trained, validated and tested on this dataset. This data set can be got accessed via tianchi.aliyun.com/competition/entrance/532036/information.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge