Junhao Ran
VisRAG: Vision-based Retrieval-augmented Generation on Multi-modality Documents
Oct 14, 2024



Abstract:Retrieval-augmented generation (RAG) is an effective technique that enables large language models (LLMs) to utilize external knowledge sources for generation. However, current RAG systems are solely based on text, rendering it impossible to utilize vision information like layout and images that play crucial roles in real-world multi-modality documents. In this paper, we introduce VisRAG, which tackles this issue by establishing a vision-language model (VLM)-based RAG pipeline. In this pipeline, instead of first parsing the document to obtain text, the document is directly embedded using a VLM as an image and then retrieved to enhance the generation of a VLM. Compared to traditional text-based RAG, VisRAG maximizes the retention and utilization of the data information in the original documents, eliminating the information loss introduced during the parsing process. We collect both open-source and synthetic data to train the retriever in VisRAG and explore a variety of generation methods. Experiments demonstrate that VisRAG outperforms traditional RAG in both the retrieval and generation stages, achieving a 25--39\% end-to-end performance gain over traditional text-based RAG pipeline. Further analysis reveals that VisRAG is effective in utilizing training data and demonstrates strong generalization capability, positioning it as a promising solution for RAG on multi-modality documents. Our code and data are available at https://github.com/openbmb/visrag .
Assessing and Enhancing Large Language Models in Rare Disease Question-answering
Aug 15, 2024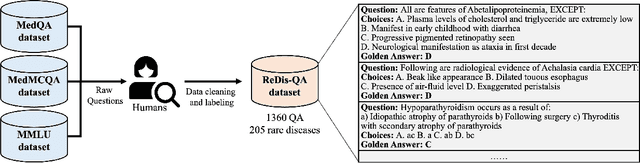

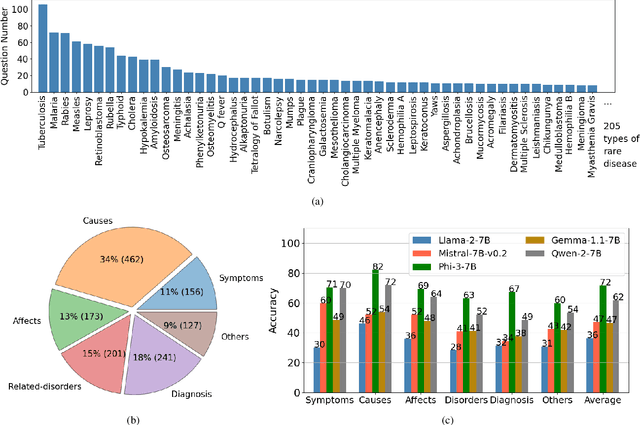
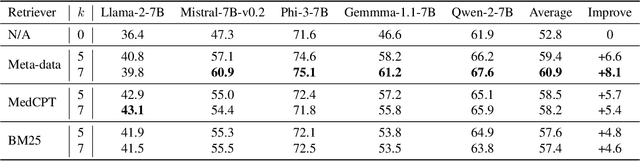
Abstract:Despite the impressive capabilities of Large Language Models (LLMs) in general medical domains, questions remain about their performance in diagnosing rare diseases. To answer this question, we aim to assess the diagnostic performance of LLMs in rare diseases, and explore methods to enhance their effectiveness in this area. In this work, we introduce a rare disease question-answering (ReDis-QA) dataset to evaluate the performance of LLMs in diagnosing rare diseases. Specifically, we collected 1360 high-quality question-answer pairs within the ReDis-QA dataset, covering 205 rare diseases. Additionally, we annotated meta-data for each question, facilitating the extraction of subsets specific to any given disease and its property. Based on the ReDis-QA dataset, we benchmarked several open-source LLMs, revealing that diagnosing rare diseases remains a significant challenge for these models. To facilitate retrieval augmentation generation for rare disease diagnosis, we collect the first rare diseases corpus (ReCOP), sourced from the National Organization for Rare Disorders (NORD) database. Specifically, we split the report of each rare disease into multiple chunks, each representing a different property of the disease, including their overview, symptoms, causes, effects, related disorders, diagnosis, and standard therapies. This structure ensures that the information within each chunk aligns consistently with a question. Experiment results demonstrate that ReCOP can effectively improve the accuracy of LLMs on the ReDis-QA dataset by an average of 8%. Moreover, it significantly guides LLMs to generate trustworthy answers and explanations that can be traced back to existing literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge