Juan Shu
Time Series Treatment Effects Analysis with Always-Missing Controls
Feb 18, 2025

Abstract:Estimating treatment effects in time series data presents a significant challenge, especially when the control group is always unobservable. For example, in analyzing the effects of Christmas on retail sales, we lack direct observation of what would have occurred in late December without the Christmas impact. To address this, we try to recover the control group in the event period while accounting for confounders and temporal dependencies. Experimental results on the M5 Walmart retail sales data demonstrate robust estimation of the potential outcome of the control group as well as accurate predicted holiday effect. Furthermore, we provided theoretical guarantees for the estimated treatment effect, proving its consistency and asymptotic normality. The proposed methodology is applicable not only to this always-missing control scenario but also in other conventional time series causal inference settings.
GAD-NR: Graph Anomaly Detection via Neighborhood Reconstruction
Jun 07, 2023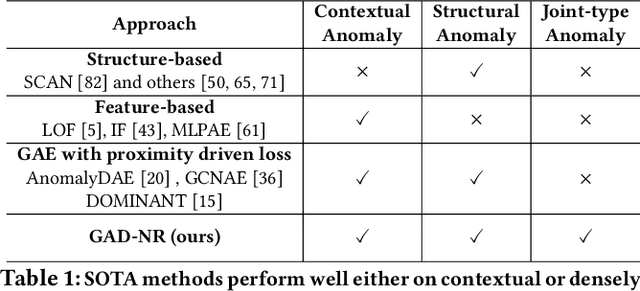


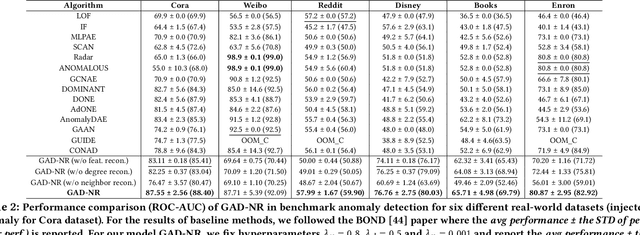
Abstract:Graph Anomaly Detection (GAD) is a technique used to identify abnormal nodes within graphs, finding applications in network security, fraud detection, social media spam detection, and various other domains. A common method for GAD is Graph Auto-Encoders (GAEs), which encode graph data into node representations and identify anomalies by assessing the reconstruction quality of the graphs based on these representations. However, existing GAE models are primarily optimized for direct link reconstruction, resulting in nodes connected in the graph being clustered in the latent space. As a result, they excel at detecting cluster-type structural anomalies but struggle with more complex structural anomalies that do not conform to clusters. To address this limitation, we propose a novel solution called GAD-NR, a new variant of GAE that incorporates neighborhood reconstruction for graph anomaly detection. GAD-NR aims to reconstruct the entire neighborhood of a node, encompassing the local structure, self-attributes, and neighbor attributes, based on the corresponding node representation. By comparing the neighborhood reconstruction loss between anomalous nodes and normal nodes, GAD-NR can effectively detect any anomalies. Extensive experimentation conducted on six real-world datasets validates the effectiveness of GAD-NR, showcasing significant improvements (by up to 30% in AUC) over state-of-the-art competitors. The source code for GAD-NR is openly available. Importantly, the comparative analysis reveals that the existing methods perform well only in detecting one or two types of anomalies out of the three types studied. In contrast, GAD-NR excels at detecting all three types of anomalies across the datasets, demonstrating its comprehensive anomaly detection capabilities.
Contrastive Cycle Adversarial Autoencoders for Single-cell Multi-omics Alignment and Integration
Dec 14, 2021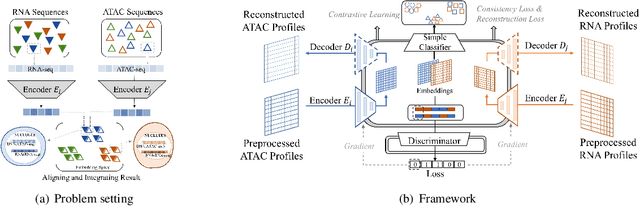
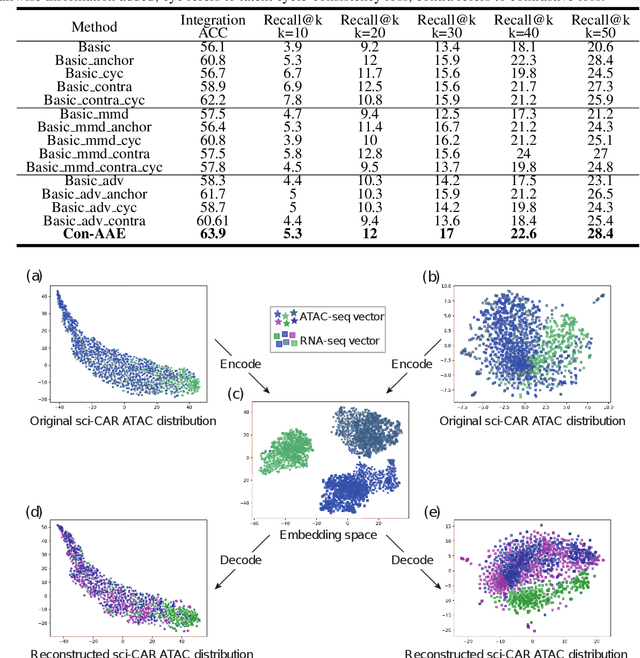
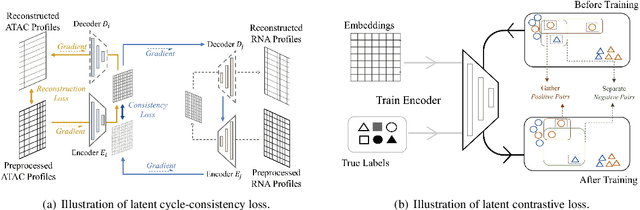
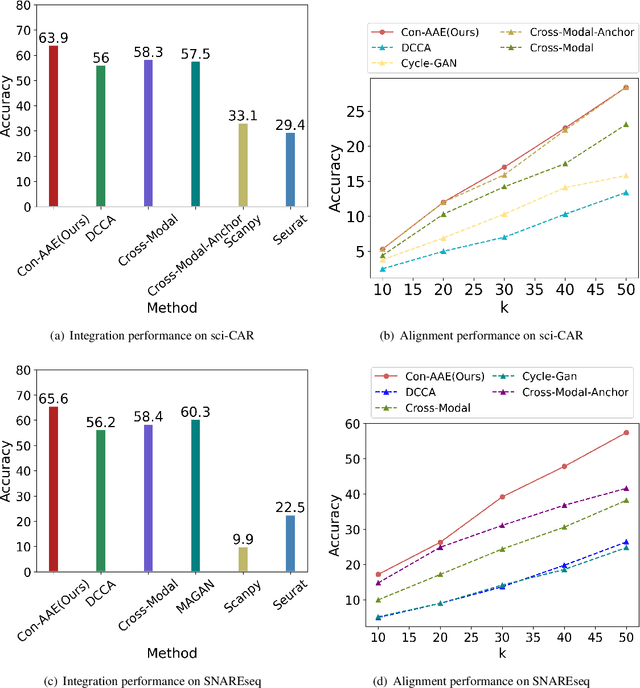
Abstract:Muilti-modality data are ubiquitous in biology, especially that we have entered the multi-omics era, when we can measure the same biological object (cell) from different aspects (omics) to provide a more comprehensive insight into the cellular system. When dealing with such multi-omics data, the first step is to determine the correspondence among different modalities. In other words, we should match data from different spaces corresponding to the same object. This problem is particularly challenging in the single-cell multi-omics scenario because such data are very sparse with extremely high dimensions. Secondly, matched single-cell multi-omics data are rare and hard to collect. Furthermore, due to the limitations of the experimental environment, the data are usually highly noisy. To promote the single-cell multi-omics research, we overcome the above challenges, proposing a novel framework to align and integrate single-cell RNA-seq data and single-cell ATAC-seq data. Our approach can efficiently map the above data with high sparsity and noise from different spaces to a low-dimensional manifold in a unified space, making the downstream alignment and integration straightforward. Compared with the other state-of-the-art methods, our method performs better in both simulated and real single-cell data. The proposed method is helpful for the single-cell multi-omics research. The improvement for integration on the simulated data is significant.
Understanding Adversarial Examples Through Deep Neural Network's Response Surface and Uncertainty Regions
Jun 30, 2021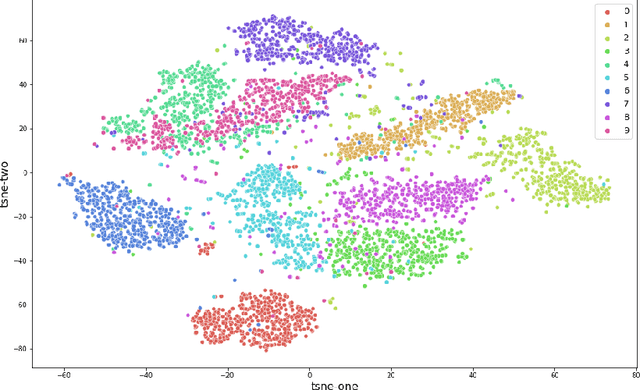

Abstract:Deep neural network (DNN) is a popular model implemented in many systems to handle complex tasks such as image classification, object recognition, natural language processing etc. Consequently DNN structural vulnerabilities become part of the security vulnerabilities in those systems. In this paper we study the root cause of DNN adversarial examples. We examine the DNN response surface to understand its classification boundary. Our study reveals the structural problem of DNN classification boundary that leads to the adversarial examples. Existing attack algorithms can generate from a handful to a few hundred adversarial examples given one clean image. We show there are infinitely many adversarial images given one clean sample, all within a small neighborhood of the clean sample. We then define DNN uncertainty regions and show transferability of adversarial examples is not universal. We also argue that generalization error, the large sample theoretical guarantee established for DNN, cannot adequately capture the phenomenon of adversarial examples. We need new theory to measure DNN robustness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge