Jonathan E. Zuckerman
Virtual histological staining of unlabeled autopsy tissue
Aug 02, 2023Abstract:Histological examination is a crucial step in an autopsy; however, the traditional histochemical staining of post-mortem samples faces multiple challenges, including the inferior staining quality due to autolysis caused by delayed fixation of cadaver tissue, as well as the resource-intensive nature of chemical staining procedures covering large tissue areas, which demand substantial labor, cost, and time. These challenges can become more pronounced during global health crises when the availability of histopathology services is limited, resulting in further delays in tissue fixation and more severe staining artifacts. Here, we report the first demonstration of virtual staining of autopsy tissue and show that a trained neural network can rapidly transform autofluorescence images of label-free autopsy tissue sections into brightfield equivalent images that match hematoxylin and eosin (H&E) stained versions of the same samples, eliminating autolysis-induced severe staining artifacts inherent in traditional histochemical staining of autopsied tissue. Our virtual H&E model was trained using >0.7 TB of image data and a data-efficient collaboration scheme that integrates the virtual staining network with an image registration network. The trained model effectively accentuated nuclear, cytoplasmic and extracellular features in new autopsy tissue samples that experienced severe autolysis, such as COVID-19 samples never seen before, where the traditional histochemical staining failed to provide consistent staining quality. This virtual autopsy staining technique can also be extended to necrotic tissue, and can rapidly and cost-effectively generate artifact-free H&E stains despite severe autolysis and cell death, also reducing labor, cost and infrastructure requirements associated with the standard histochemical staining.
Deep learning-based transformation of the H&E stain into special stains improves kidney disease diagnosis
Aug 20, 2020
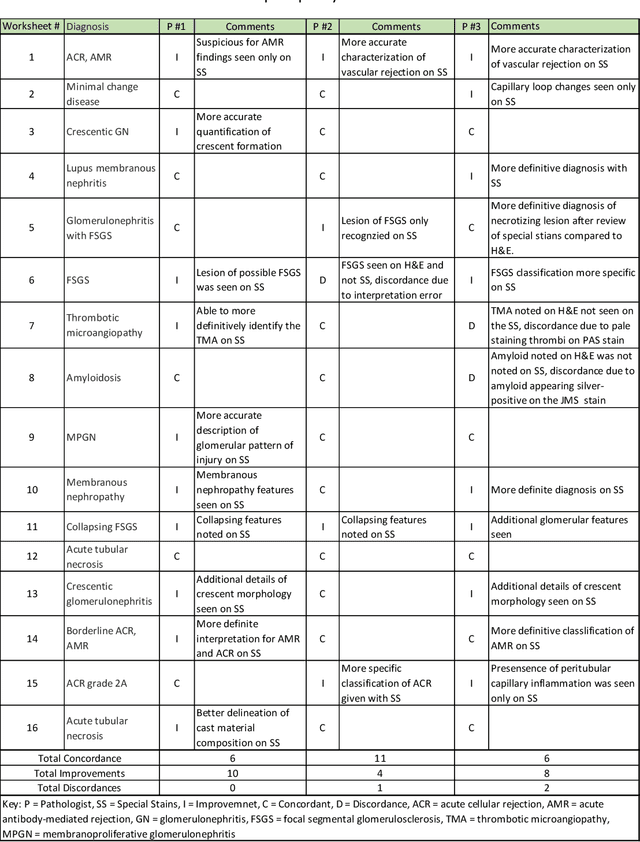


Abstract:Pathology is practiced by visual inspection of histochemically stained slides. Most commonly, the hematoxylin and eosin (H&E) stain is used in the diagnostic workflow and it is the gold standard for cancer diagnosis. However, in many cases, especially for non-neoplastic diseases, additional "special stains" are used to provide different levels of contrast and color to tissue components and allow pathologists to get a clearer diagnostic picture. In this study, we demonstrate the utility of supervised learning-based computational stain transformation from H&E to different special stains (Masson's Trichrome, periodic acid-Schiff and Jones silver stain) using tissue sections from kidney needle core biopsies. Based on evaluation by three renal pathologists, followed by adjudication by a fourth renal pathologist, we show that the generation of virtual special stains from existing H&E images improves the diagnosis in several non-neoplastic kidney diseases, sampled from 16 unique subjects. Adjudication of N=48 diagnoses from the three pathologists revealed that the virtually generated special stains yielded 22 improvements (45.8%), 23 concordances (47.9%) and 3 discordances (6.3%), when compared against the use of H&E stained tissue only. As the virtual transformation of H&E images into special stains can be achieved in less than 1 min per patient core specimen slide, this stain-to-stain transformation framework can improve the quality of the preliminary diagnosis when additional special stains are needed, along with significant savings in time and cost, reducing the burden on healthcare system and patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge