Jiancong Wang
for the Alzheimer's Disease Neuroimaging Initiative
Regional Deep Atrophy: a Self-Supervised Learning Method to Automatically Identify Regions Associated With Alzheimer's Disease Progression From Longitudinal MRI
Apr 10, 2023Abstract:Longitudinal assessment of brain atrophy, particularly in the hippocampus, is a well-studied biomarker for neurodegenerative diseases, such as Alzheimer's disease (AD). In clinical trials, estimation of brain progressive rates can be applied to track therapeutic efficacy of disease modifying treatments. However, most state-of-the-art measurements calculate changes directly by segmentation and/or deformable registration of MRI images, and may misreport head motion or MRI artifacts as neurodegeneration, impacting their accuracy. In our previous study, we developed a deep learning method DeepAtrophy that uses a convolutional neural network to quantify differences between longitudinal MRI scan pairs that are associated with time. DeepAtrophy has high accuracy in inferring temporal information from longitudinal MRI scans, such as temporal order or relative inter-scan interval. DeepAtrophy also provides an overall atrophy score that was shown to perform well as a potential biomarker of disease progression and treatment efficacy. However, DeepAtrophy is not interpretable, and it is unclear what changes in the MRI contribute to progression measurements. In this paper, we propose Regional Deep Atrophy (RDA), which combines the temporal inference approach from DeepAtrophy with a deformable registration neural network and attention mechanism that highlights regions in the MRI image where longitudinal changes are contributing to temporal inference. RDA has similar prediction accuracy as DeepAtrophy, but its additional interpretability makes it more acceptable for use in clinical settings, and may lead to more sensitive biomarkers for disease monitoring in clinical trials of early AD.
Deformable Image Registration using Neural ODEs
Aug 27, 2021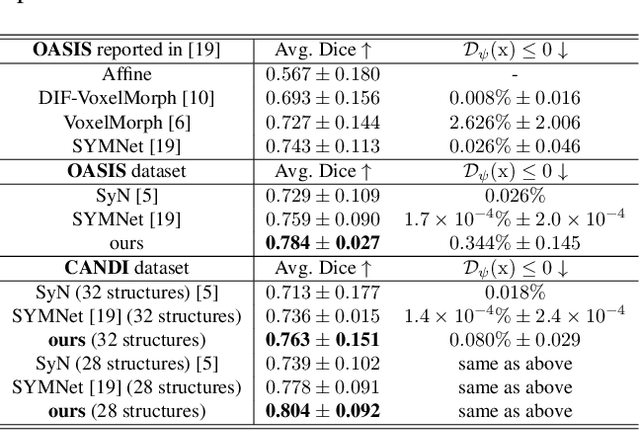
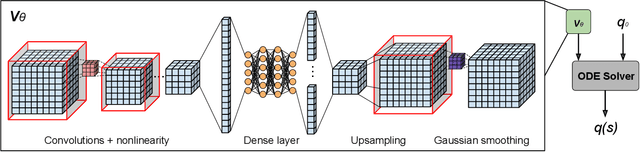
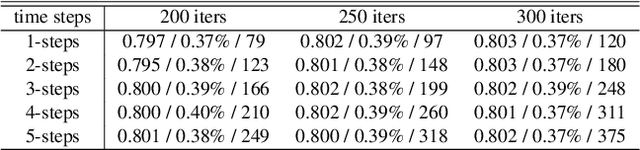
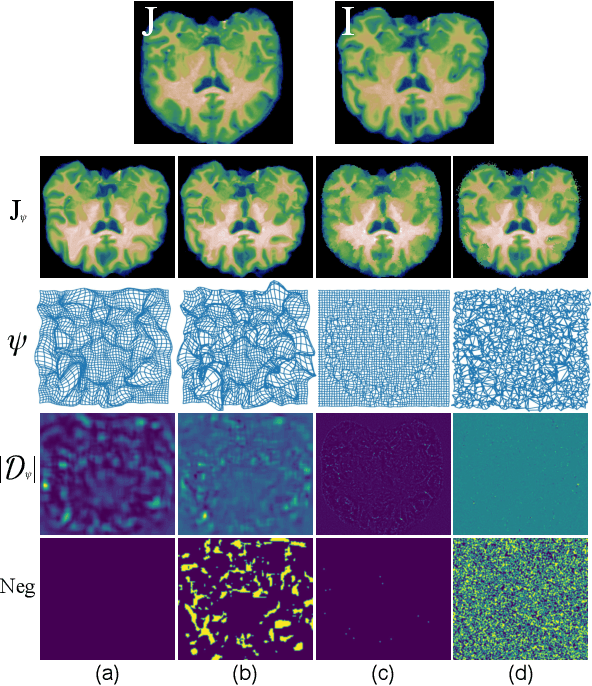
Abstract:Deformable image registration, aiming to find spatial correspondence between a given image pair, is one of the most critical problems in the domain of medical image analysis. In this paper, we present a generic, fast, and accurate diffeomorphic image registration framework that leverages neural ordinary differential equations (NODEs). We model each voxel as a moving particle and consider the set of all voxels in a 3D image as a high-dimensional dynamical system whose trajectory determines the targeted deformation field. Compared with traditional optimization-based methods, our framework reduces the running time from tens of minutes to tens of seconds. Compared with recent data-driven deep learning methods, our framework is more accessible since it does not require large amounts of training data. Our experiments show that the registration results of our method outperform state-of-the-arts under various metrics, indicating that our modeling approach is well fitted for the task of deformable image registration.
Deep Label Fusion: A 3D End-to-End Hybrid Multi-Atlas Segmentation and Deep Learning Pipeline
Mar 19, 2021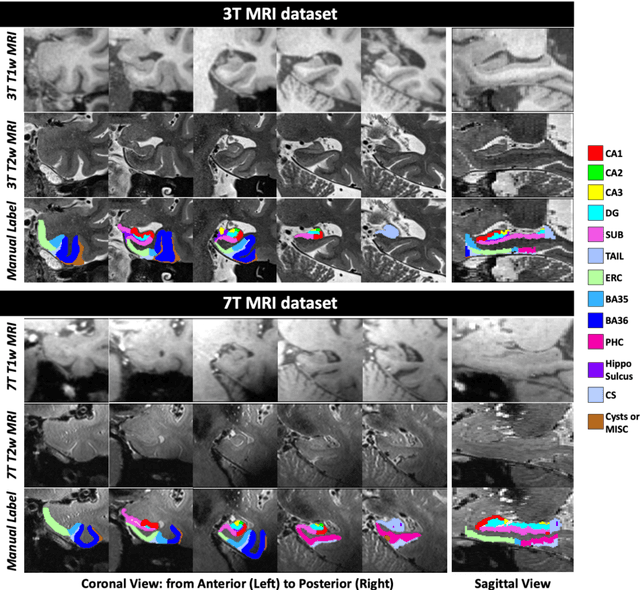
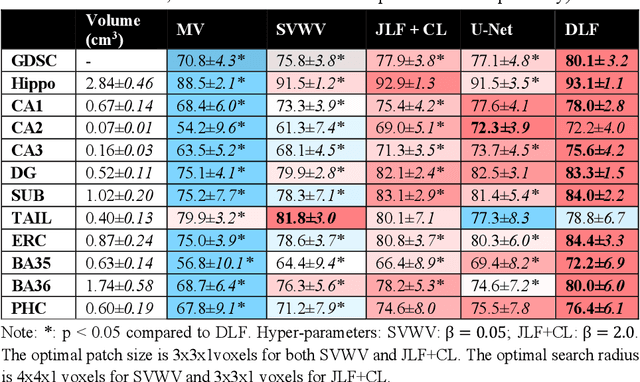
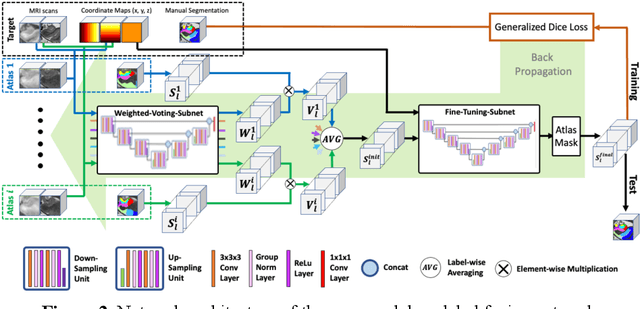

Abstract:Deep learning (DL) is the state-of-the-art methodology in various medical image segmentation tasks. However, it requires relatively large amounts of manually labeled training data, which may be infeasible to generate in some applications. In addition, DL methods have relatively poor generalizability to out-of-sample data. Multi-atlas segmentation (MAS), on the other hand, has promising performance using limited amounts of training data and good generalizability. A hybrid method that integrates the high accuracy of DL and good generalizability of MAS is highly desired and could play an important role in segmentation problems where manually labeled data is hard to generate. Most of the prior work focuses on improving single components of MAS using DL rather than directly optimizing the final segmentation accuracy via an end-to-end pipeline. Only one study explored this idea in binary segmentation of 2D images, but it remains unknown whether it generalizes well to multi-class 3D segmentation problems. In this study, we propose a 3D end-to-end hybrid pipeline, named deep label fusion (DLF), that takes advantage of the strengths of MAS and DL. Experimental results demonstrate that DLF yields significant improvements over conventional label fusion methods and U-Net, a direct DL approach, in the context of segmenting medial temporal lobe subregions using 3T T1-weighted and T2-weighted MRI. Further, when applied to an unseen similar dataset acquired in 7T, DLF maintains its superior performance, which demonstrates its good generalizability.
DeepAtrophy: Teaching a Neural Network to Differentiate Progressive Changes from Noise on Longitudinal MRI in Alzheimer's Disease
Oct 24, 2020



Abstract:Volume change measures derived from longitudinal MRI (e.g. hippocampal atrophy) are a well-studied biomarker of disease progression in Alzheimer's Disease (AD) and are used in clinical trials to track the therapeutic efficacy of disease-modifying treatments. However, longitudinal MRI change measures can be confounded by non-biological factors, such as different degrees of head motion and susceptibility artifact between pairs of MRI scans. We hypothesize that deep learning methods applied directly to pairs of longitudinal MRI scans can be trained to differentiate between biological changes and non-biological factors better than conventional approaches based on deformable image registration. To achieve this, we make a simplifying assumption that biological factors are associated with time (i.e. the hippocampus shrinks overtime in the aging population) whereas non-biological factors are independent of time. We then formulate deep learning networks to infer the temporal order of same-subject MRI scans input to the network in arbitrary order; as well as to infer ratios between interscan intervals for two pairs of same-subject MRI scans. In the test dataset, these networks perform better in tasks of temporal ordering (89.3%) and interscan interval inference (86.1%) than a state-of-the-art deformation-based morphometry method ALOHA (76.6% and 76.1% respectively) (Das et al., 2012). Furthermore, we derive a disease progression score from the network that is able to detect a group difference between 58 preclinical AD and 75 beta-amyloid-negative cognitively normal individuals within one year, compared to two years for ALOHA. This suggests that deep learning can be trained to differentiate MRI changes due to biological factors (tissue loss) from changes due to non-biological factors, leading to novel biomarkers that are more sensitive to longitudinal changes at the earliest stages of AD.
Nested Scale Editing for Conditional Image Synthesis
Jun 03, 2020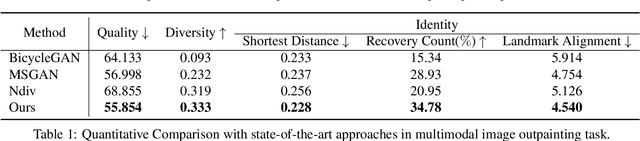
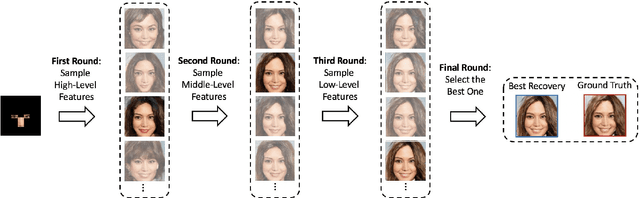
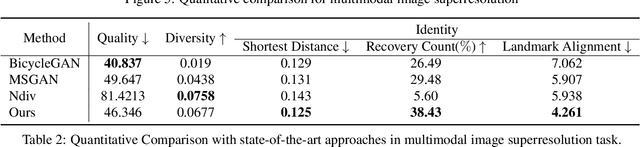
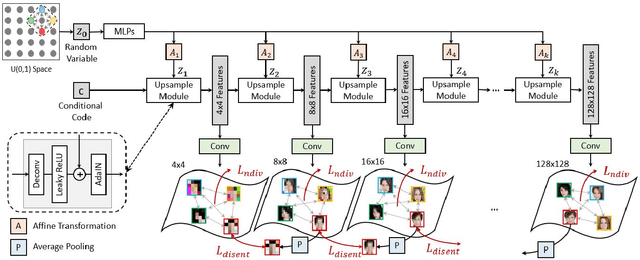
Abstract:We propose an image synthesis approach that provides stratified navigation in the latent code space. With a tiny amount of partial or very low-resolution image, our approach can consistently out-perform state-of-the-art counterparts in terms of generating the closest sampled image to the ground truth. We achieve this through scale-independent editing while expanding scale-specific diversity. Scale-independence is achieved with a nested scale disentanglement loss. Scale-specific diversity is created by incorporating a progressive diversification constraint. We introduce semantic persistency across the scales by sharing common latent codes. Together they provide better control of the image synthesis process. We evaluate the effectiveness of our proposed approach through various tasks, including image outpainting, image superresolution, and cross-domain image translation.
Multimodal Image Outpainting With Regularized Normalized Diversification
Oct 25, 2019



Abstract:In this paper, we study the problem of generating a set ofrealistic and diverse backgrounds when given only a smallforeground region. We refer to this task as image outpaint-ing. The technical challenge of this task is to synthesize notonly plausible but also diverse image outputs. Traditionalgenerative adversarial networks suffer from mode collapse.While recent approaches propose to maximize orpreserve the pairwise distance between generated sampleswith respect to their latent distance, they do not explicitlyprevent the diverse samples of different conditional inputsfrom collapsing. Therefore, we propose a new regulariza-tion method to encourage diverse sampling in conditionalsynthesis. In addition, we propose a feature pyramid dis-criminator to improve the image quality. Our experimen-tal results show that our model can produce more diverseimages without sacrificing visual quality compared to state-of-the-arts approaches in both the CelebA face dataset and the Cityscape scene dataset.
Enhanced generative adversarial network for 3D brain MRI super-resolution
Jul 15, 2019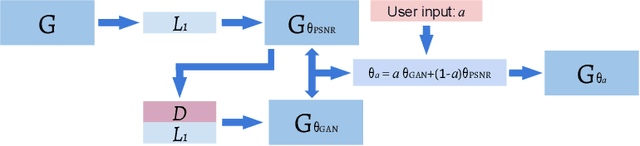
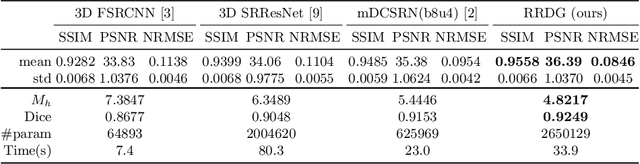
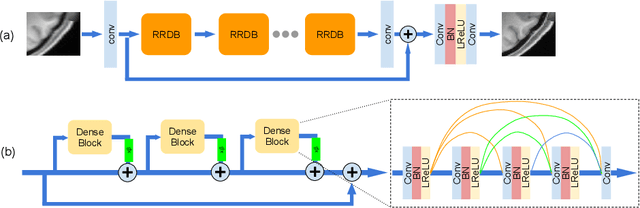
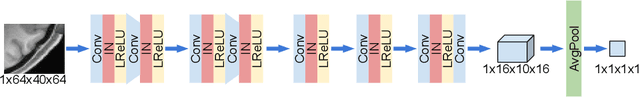
Abstract:Single image super-resolution (SISR) reconstruction for magnetic resonance imaging (MRI) has generated significant interest because of its potential to not only speed up imaging but to improve quantitative processing and analysis of available image data. Generative Adversarial Networks (GAN) have proven to perform well in recovering image texture detail, and many variants have therefore been proposed for SISR. In this work, we develop an enhancement to tackle GAN-based 3D SISR by introducing a new residual-in-residual dense block (RRDG) generator that is both memory efficient and achieves state-of-the-art performance in terms of PSNR (Peak Signal to Noise Ratio), SSIM (Structural Similarity) and NRMSE (Normalized Root Mean Squared Error) metrics. We also introduce a patch GAN discriminator with improved convergence behavior to better model brain image texture. We proposed a novel the anatomical fidelity evaluation of the results using a pre-trained brain parcellation network. Finally, these developments are combined through a simple and efficient method to balance etween image and texture quality in the final output.
Fast geodesic shooting for landmark matching using CUDA
Jul 10, 2019


Abstract:Landmark matching via geodesic shooting is a prerequisite task for numerous registration based applications in biomedicine. Geodesic shooting has been developed as one solution approach and formulates the diffeomorphic registration as an optimal control problem under the Hamiltonian framework. In this framework, with landmark positions q0 fixed, the problem solely depends on the initial momentum p0 and evolves through time steps according to a set of constraint equations. Given an initial p0, the algorithm flows q and p forward through time steps, calculates a loss based on point-set mismatch and kinetic energy, back-propagate through time to calculate gradient on p0 and update it. In the forward and backward pass, a pair-wise kernel on landmark points K and additional intermediate terms have to be calculated and marginalized, leading to O(N2) computational complexity, N being the number of points to be registered. For medical image applications, N maybe in the range of thousands, rendering this operation computationally expensive. In this work we ropose a CUDA implementation based on shared memory reduction. Our implementation achieves nearly 2 orders magnitude speed up compared to a naive CPU-based implementation, in addition to improved numerical accuracy as well as better registration results.
Barnes-Hut Approximation for Point SetGeodesic Shooting
Jul 10, 2019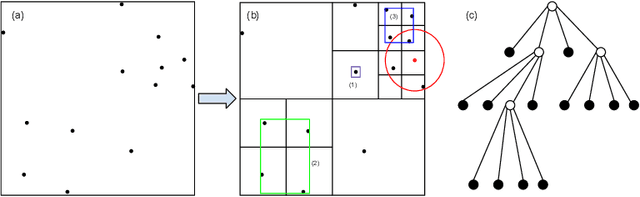

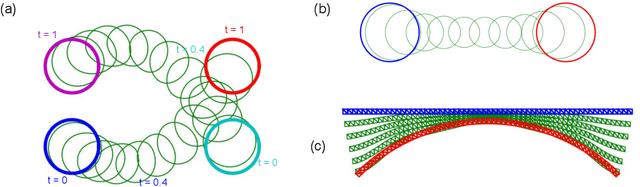

Abstract:Geodesic shooting has been successfully applied to diffeo-morphic registration of point sets. Exact computation of the geodesicshooting between point sets, however, requiresO(N2) calculations each time step on the number of points in the point set. We proposean approximation approach based on the Barnes-Hut algorithm to speedup point set geodesic shooting. This approximation can reduce the al-gorithm complexity toO(N b+N logN). The evaluation of the proposedmethod in both simulated images and the medial temporal lobe thick-ness analysis demonstrates a comparable accuracy to the exact point set geodesic shooting while offering up to 3-fold speed up. This improvementopens up a range of clinical research studies and practical problems towhich the method can be effectively applied.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge