Sandhitsu R. Das
for the Alzheimer's Disease Neuroimaging Initiative
Development of an isotropic segmentation model for medial temporal lobe subregions on anisotropic MRI atlas using implicit neural representation
Aug 24, 2025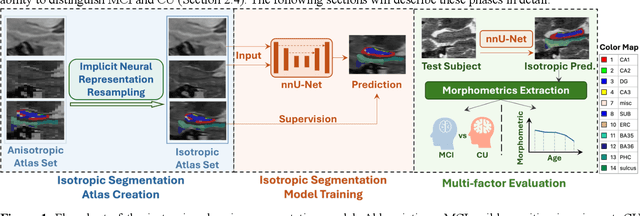

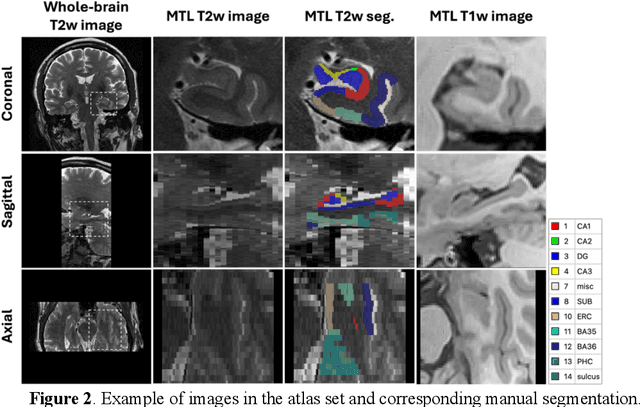
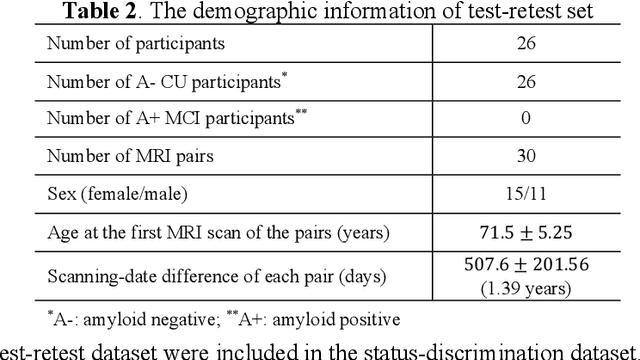
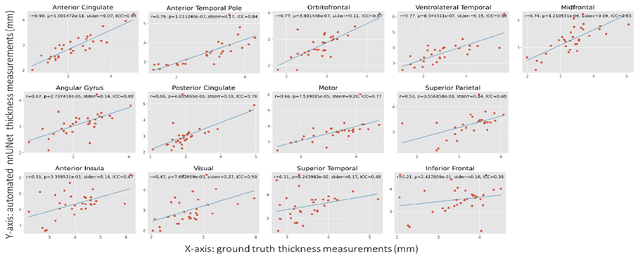
Abstract:Imaging biomarkers in magnetic resonance imaging (MRI) are important tools for diagnosing and tracking Alzheimer's disease (AD). As medial temporal lobe (MTL) is the earliest region to show AD-related hallmarks, brain atrophy caused by AD can first be observed in the MTL. Accurate segmentation of MTL subregions and extraction of imaging biomarkers from them are important. However, due to imaging limitations, the resolution of T2-weighted (T2w) MRI is anisotropic, which makes it difficult to accurately extract the thickness of cortical subregions in the MTL. In this study, we used an implicit neural representation method to combine the resolution advantages of T1-weighted and T2w MRI to accurately upsample an MTL subregion atlas set from anisotropic space to isotropic space, establishing a multi-modality, high-resolution atlas set. Based on this atlas, we developed an isotropic MTL subregion segmentation model. In an independent test set, the cortical subregion thickness extracted using this isotropic model showed higher significance than an anisotropic method in distinguishing between participants with mild cognitive impairment and cognitively unimpaired (CU) participants. In longitudinal analysis, the biomarkers extracted using isotropic method showed greater stability in CU participants. This study improved the accuracy of AD imaging biomarkers without increasing the amount of atlas annotation work, which may help to more accurately quantify the relationship between AD and brain atrophy and provide more accurate measures for disease tracking.
Nearly isotropic segmentation for medial temporal lobe subregions in multi-modality MRI
Apr 25, 2025Abstract:Morphometry of medial temporal lobe (MTL) subregions in brain MRI is sensitive biomarker to Alzheimers Disease and other related conditions. While T2-weighted (T2w) MRI with high in-plane resolution is widely used to segment hippocampal subfields due to its higher contrast in hippocampus, its lower out-of-plane resolution reduces the accuracy of subregion thickness measurements. To address this issue, we developed a nearly isotropic segmentation pipeline that incorporates image and label upsampling and high-resolution segmentation in T2w MRI. First, a high-resolution atlas was created based on an existing anisotropic atlas derived from 29 individuals. Both T1-weighted and T2w images in the atlas were upsampled from their original resolution to a nearly isotropic resolution 0.4x0.4x0.52mm3 using a non-local means approach. Manual segmentations within the atlas were also upsampled to match this resolution using a UNet-based neural network, which was trained on a cohort consisting of both high-resolution ex vivo and low-resolution anisotropic in vivo MRI with manual segmentations. Second, a multi-modality deep learning-based segmentation model was trained within this nearly isotropic atlas. Finally, experiments showed the nearly isotropic subregion segmentation improved the accuracy of cortical thickness as an imaging biomarker for neurodegeneration in T2w MRI.
Regional Deep Atrophy: a Self-Supervised Learning Method to Automatically Identify Regions Associated With Alzheimer's Disease Progression From Longitudinal MRI
Apr 10, 2023Abstract:Longitudinal assessment of brain atrophy, particularly in the hippocampus, is a well-studied biomarker for neurodegenerative diseases, such as Alzheimer's disease (AD). In clinical trials, estimation of brain progressive rates can be applied to track therapeutic efficacy of disease modifying treatments. However, most state-of-the-art measurements calculate changes directly by segmentation and/or deformable registration of MRI images, and may misreport head motion or MRI artifacts as neurodegeneration, impacting their accuracy. In our previous study, we developed a deep learning method DeepAtrophy that uses a convolutional neural network to quantify differences between longitudinal MRI scan pairs that are associated with time. DeepAtrophy has high accuracy in inferring temporal information from longitudinal MRI scans, such as temporal order or relative inter-scan interval. DeepAtrophy also provides an overall atrophy score that was shown to perform well as a potential biomarker of disease progression and treatment efficacy. However, DeepAtrophy is not interpretable, and it is unclear what changes in the MRI contribute to progression measurements. In this paper, we propose Regional Deep Atrophy (RDA), which combines the temporal inference approach from DeepAtrophy with a deformable registration neural network and attention mechanism that highlights regions in the MRI image where longitudinal changes are contributing to temporal inference. RDA has similar prediction accuracy as DeepAtrophy, but its additional interpretability makes it more acceptable for use in clinical settings, and may lead to more sensitive biomarkers for disease monitoring in clinical trials of early AD.
Automated deep learning segmentation of high-resolution 7 T ex vivo MRI for quantitative analysis of structure-pathology correlations in neurodegenerative diseases
Mar 21, 2023



Abstract:Ex vivo MRI of the brain provides remarkable advantages over in vivo MRI for visualizing and characterizing detailed neuroanatomy, and helps to link microscale histology studies with morphometric measurements. However, automated segmentation methods for brain mapping in ex vivo MRI are not well developed, primarily due to limited availability of labeled datasets, and heterogeneity in scanner hardware and acquisition protocols. In this work, we present a high resolution dataset of 37 ex vivo post-mortem human brain tissue specimens scanned on a 7T whole-body MRI scanner. We developed a deep learning pipeline to segment the cortical mantle by benchmarking the performance of nine deep neural architectures. We then segment the four subcortical structures: caudate, putamen, globus pallidus, and thalamus; white matter hyperintensities, and the normal appearing white matter. We show excellent generalizing capabilities across whole brain hemispheres in different specimens, and also on unseen images acquired at different magnetic field strengths and different imaging sequence. We then compute volumetric and localized cortical thickness measurements across key regions, and link them with semi-quantitative neuropathological ratings. Our code, containerized executables, and the processed datasets are publicly available at: https://github.com/Pulkit-Khandelwal/upenn-picsl-brain-ex-vivo.
Gray Matter Segmentation in Ultra High Resolution 7 Tesla ex vivo T2w MRI of Human Brain Hemispheres
Oct 14, 2021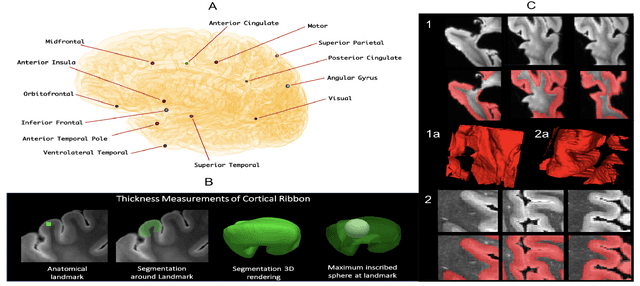
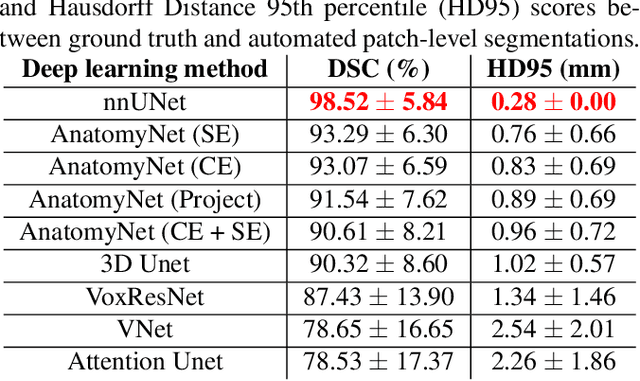
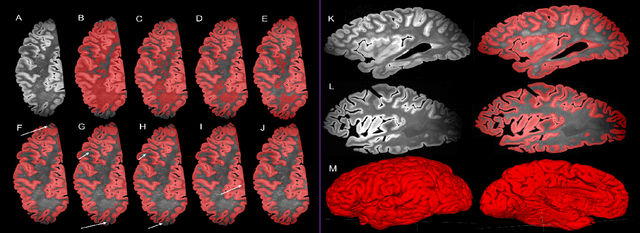
Abstract:Ex vivo MRI of the brain provides remarkable advantages over in vivo MRI for visualizing and characterizing detailed neuroanatomy. However, automated cortical segmentation methods in ex vivo MRI are not well developed, primarily due to limited availability of labeled datasets, and heterogeneity in scanner hardware and acquisition protocols. In this work, we present a high resolution 7 Tesla dataset of 32 ex vivo human brain specimens. We benchmark the cortical mantle segmentation performance of nine neural network architectures, trained and evaluated using manually-segmented 3D patches sampled from specific cortical regions, and show excellent generalizing capabilities across whole brain hemispheres in different specimens, and also on unseen images acquired at different magnetic field strength and imaging sequences. Finally, we provide cortical thickness measurements across key regions in 3D ex vivo human brain images. Our code and processed datasets are publicly available at https://github.com/Pulkit-Khandelwal/picsl-ex-vivo-segmentation.
DeepAtrophy: Teaching a Neural Network to Differentiate Progressive Changes from Noise on Longitudinal MRI in Alzheimer's Disease
Oct 24, 2020



Abstract:Volume change measures derived from longitudinal MRI (e.g. hippocampal atrophy) are a well-studied biomarker of disease progression in Alzheimer's Disease (AD) and are used in clinical trials to track the therapeutic efficacy of disease-modifying treatments. However, longitudinal MRI change measures can be confounded by non-biological factors, such as different degrees of head motion and susceptibility artifact between pairs of MRI scans. We hypothesize that deep learning methods applied directly to pairs of longitudinal MRI scans can be trained to differentiate between biological changes and non-biological factors better than conventional approaches based on deformable image registration. To achieve this, we make a simplifying assumption that biological factors are associated with time (i.e. the hippocampus shrinks overtime in the aging population) whereas non-biological factors are independent of time. We then formulate deep learning networks to infer the temporal order of same-subject MRI scans input to the network in arbitrary order; as well as to infer ratios between interscan intervals for two pairs of same-subject MRI scans. In the test dataset, these networks perform better in tasks of temporal ordering (89.3%) and interscan interval inference (86.1%) than a state-of-the-art deformation-based morphometry method ALOHA (76.6% and 76.1% respectively) (Das et al., 2012). Furthermore, we derive a disease progression score from the network that is able to detect a group difference between 58 preclinical AD and 75 beta-amyloid-negative cognitively normal individuals within one year, compared to two years for ALOHA. This suggests that deep learning can be trained to differentiate MRI changes due to biological factors (tissue loss) from changes due to non-biological factors, leading to novel biomarkers that are more sensitive to longitudinal changes at the earliest stages of AD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge