Paul Yushkevich
for the Alzheimer's Disease Neuroimaging Initiative
DeepAtrophy: Teaching a Neural Network to Differentiate Progressive Changes from Noise on Longitudinal MRI in Alzheimer's Disease
Oct 24, 2020



Abstract:Volume change measures derived from longitudinal MRI (e.g. hippocampal atrophy) are a well-studied biomarker of disease progression in Alzheimer's Disease (AD) and are used in clinical trials to track the therapeutic efficacy of disease-modifying treatments. However, longitudinal MRI change measures can be confounded by non-biological factors, such as different degrees of head motion and susceptibility artifact between pairs of MRI scans. We hypothesize that deep learning methods applied directly to pairs of longitudinal MRI scans can be trained to differentiate between biological changes and non-biological factors better than conventional approaches based on deformable image registration. To achieve this, we make a simplifying assumption that biological factors are associated with time (i.e. the hippocampus shrinks overtime in the aging population) whereas non-biological factors are independent of time. We then formulate deep learning networks to infer the temporal order of same-subject MRI scans input to the network in arbitrary order; as well as to infer ratios between interscan intervals for two pairs of same-subject MRI scans. In the test dataset, these networks perform better in tasks of temporal ordering (89.3%) and interscan interval inference (86.1%) than a state-of-the-art deformation-based morphometry method ALOHA (76.6% and 76.1% respectively) (Das et al., 2012). Furthermore, we derive a disease progression score from the network that is able to detect a group difference between 58 preclinical AD and 75 beta-amyloid-negative cognitively normal individuals within one year, compared to two years for ALOHA. This suggests that deep learning can be trained to differentiate MRI changes due to biological factors (tissue loss) from changes due to non-biological factors, leading to novel biomarkers that are more sensitive to longitudinal changes at the earliest stages of AD.
Domain Generalizer: A Few-shot Meta Learning Framework for Domain Generalization in Medical Imaging
Aug 18, 2020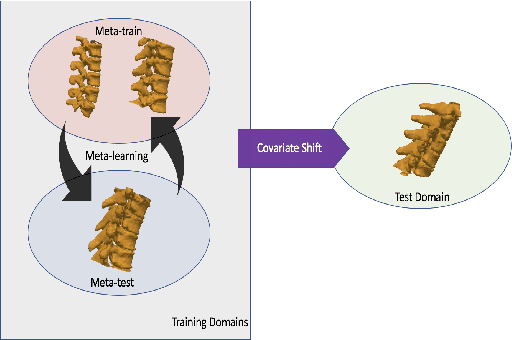
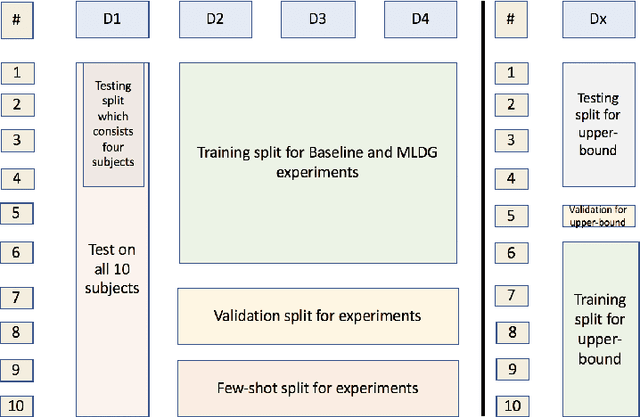

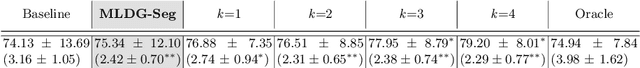
Abstract:Deep learning models perform best when tested on target (test) data domains whose distribution is similar to the set of source (train) domains. However, model generalization can be hindered when there is significant difference in the underlying statistics between the target and source domains. In this work, we adapt a domain generalization method based on a model-agnostic meta-learning framework to biomedical imaging. The method learns a domain-agnostic feature representation to improve generalization of models to the unseen test distribution. The method can be used for any imaging task, as it does not depend on the underlying model architecture. We validate the approach through a computed tomography (CT) vertebrae segmentation task across healthy and pathological cases on three datasets. Next, we employ few-shot learning, i.e. training the generalized model using very few examples from the unseen domain, to quickly adapt the model to new unseen data distribution. Our results suggest that the method could help generalize models across different medical centers, image acquisition protocols, anatomies, different regions in a given scan, healthy and diseased populations across varied imaging modalities.
Barnes-Hut Approximation for Point SetGeodesic Shooting
Jul 10, 2019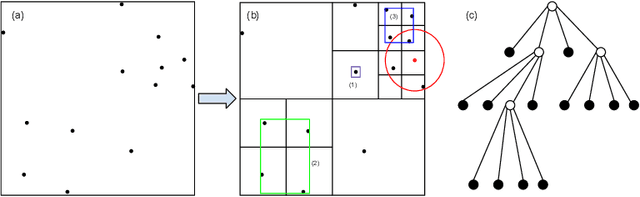

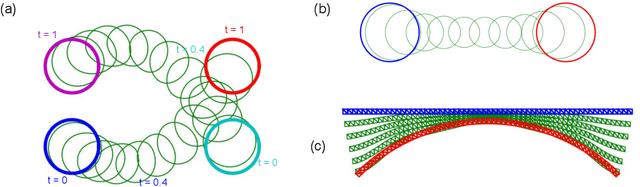

Abstract:Geodesic shooting has been successfully applied to diffeo-morphic registration of point sets. Exact computation of the geodesicshooting between point sets, however, requiresO(N2) calculations each time step on the number of points in the point set. We proposean approximation approach based on the Barnes-Hut algorithm to speedup point set geodesic shooting. This approximation can reduce the al-gorithm complexity toO(N b+N logN). The evaluation of the proposedmethod in both simulated images and the medial temporal lobe thick-ness analysis demonstrates a comparable accuracy to the exact point set geodesic shooting while offering up to 3-fold speed up. This improvementopens up a range of clinical research studies and practical problems towhich the method can be effectively applied.
Accurate and Robust Alignment of Variable-stained Histologic Images Using a General-purpose Greedy Diffeomorphic Registration Tool
Apr 26, 2019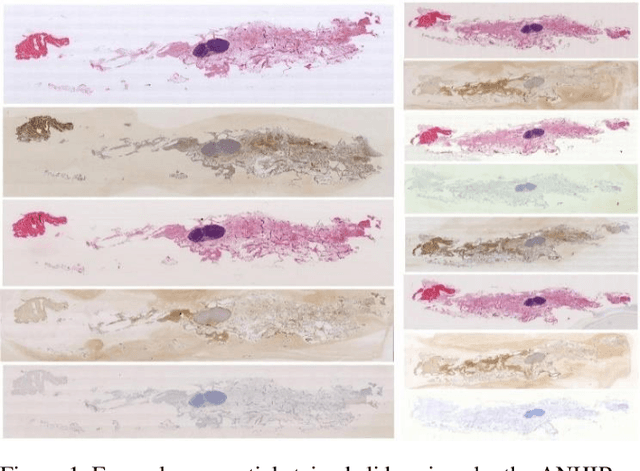
Abstract:Variously stained histology slices are routinely used by pathologists to assess extracted tissue samples from various anatomical sites and determine the presence or extent of a disease. Evaluation of sequential slides is expected to enable a better understanding of the spatial arrangement and growth patterns of cells and vessels. In this paper we present a practical two-step approach based on diffeomorphic registration to align digitized sequential histopathology stained slides to each other, starting with an initial affine step followed by the estimation of a detailed deformation field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge