Jian-Zhe Wang
MEDFuse: Multimodal EHR Data Fusion with Masked Lab-Test Modeling and Large Language Models
Jul 17, 2024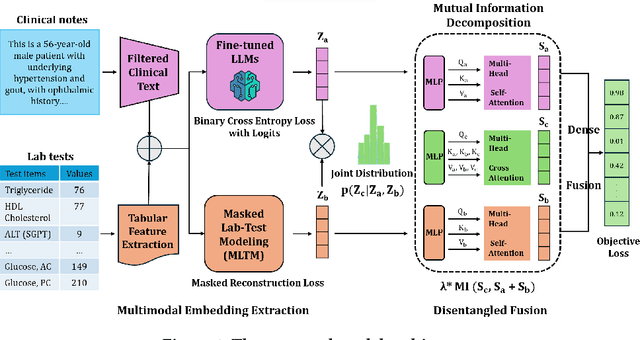
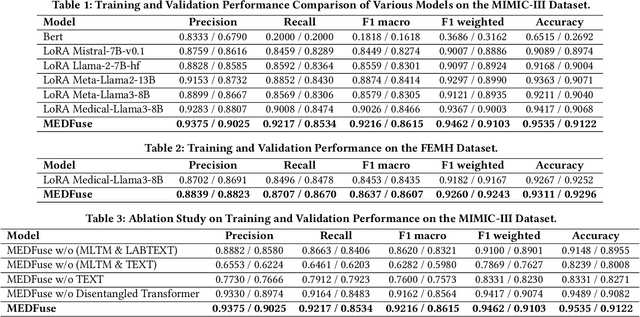
Abstract:Electronic health records (EHRs) are multimodal by nature, consisting of structured tabular features like lab tests and unstructured clinical notes. In real-life clinical practice, doctors use complementary multimodal EHR data sources to get a clearer picture of patients' health and support clinical decision-making. However, most EHR predictive models do not reflect these procedures, as they either focus on a single modality or overlook the inter-modality interactions/redundancy. In this work, we propose MEDFuse, a Multimodal EHR Data Fusion framework that incorporates masked lab-test modeling and large language models (LLMs) to effectively integrate structured and unstructured medical data. MEDFuse leverages multimodal embeddings extracted from two sources: LLMs fine-tuned on free clinical text and masked tabular transformers trained on structured lab test results. We design a disentangled transformer module, optimized by a mutual information loss to 1) decouple modality-specific and modality-shared information and 2) extract useful joint representation from the noise and redundancy present in clinical notes. Through comprehensive validation on the public MIMIC-III dataset and the in-house FEMH dataset, MEDFuse demonstrates great potential in advancing clinical predictions, achieving over 90% F1 score in the 10-disease multi-label classification task.
Large Language Multimodal Models for 5-Year Chronic Disease Cohort Prediction Using EHR Data
Mar 02, 2024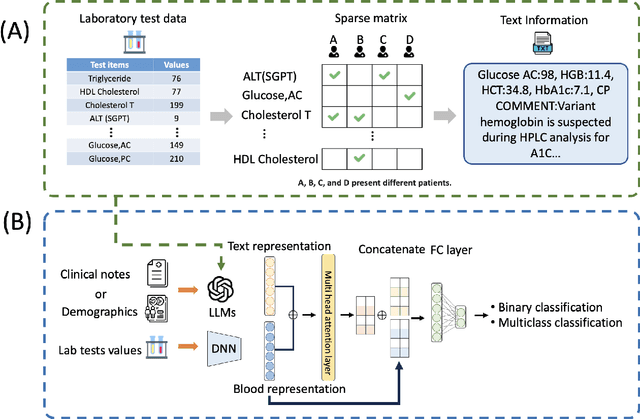
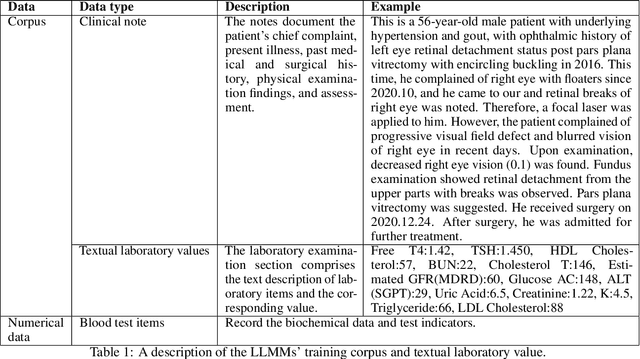

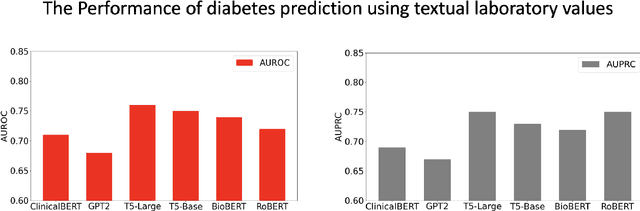
Abstract:Chronic diseases such as diabetes are the leading causes of morbidity and mortality worldwide. Numerous research studies have been attempted with various deep learning models in diagnosis. However, most previous studies had certain limitations, including using publicly available datasets (e.g. MIMIC), and imbalanced data. In this study, we collected five-year electronic health records (EHRs) from the Taiwan hospital database, including 1,420,596 clinical notes, 387,392 laboratory test results, and more than 1,505 laboratory test items, focusing on research pre-training large language models. We proposed a novel Large Language Multimodal Models (LLMMs) framework incorporating multimodal data from clinical notes and laboratory test results for the prediction of chronic disease risk. Our method combined a text embedding encoder and multi-head attention layer to learn laboratory test values, utilizing a deep neural network (DNN) module to merge blood features with chronic disease semantics into a latent space. In our experiments, we observe that clinicalBERT and PubMed-BERT, when combined with attention fusion, can achieve an accuracy of 73% in multiclass chronic diseases and diabetes prediction. By transforming laboratory test values into textual descriptions and employing the Flan T-5 model, we achieved a 76% Area Under the ROC Curve (AUROC), demonstrating the effectiveness of leveraging numerical text data for training and inference in language models. This approach significantly improves the accuracy of early-stage diabetes prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge