Hiroshi Kajino
Improving Molecular Properties Prediction Through Latent Space Fusion
Oct 20, 2023Abstract:Pre-trained Language Models have emerged as promising tools for predicting molecular properties, yet their development is in its early stages, necessitating further research to enhance their efficacy and address challenges such as generalization and sample efficiency. In this paper, we present a multi-view approach that combines latent spaces derived from state-of-the-art chemical models. Our approach relies on two pivotal elements: the embeddings derived from MHG-GNN, which represent molecular structures as graphs, and MoLFormer embeddings rooted in chemical language. The attention mechanism of MoLFormer is able to identify relations between two atoms even when their distance is far apart, while the GNN of MHG-GNN can more precisely capture relations among multiple atoms closely located. In this work, we demonstrate the superior performance of our proposed multi-view approach compared to existing state-of-the-art methods, including MoLFormer-XL, which was trained on 1.1 billion molecules, particularly in intricate tasks such as predicting clinical trial drug toxicity and inhibiting HIV replication. We assessed our approach using six benchmark datasets from MoleculeNet, where it outperformed competitors in five of them. Our study highlights the potential of latent space fusion and feature integration for advancing molecular property prediction. In this work, we use small versions of MHG-GNN and MoLFormer, which opens up an opportunity for further improvement when our approach uses a larger-scale dataset.
MHG-GNN: Combination of Molecular Hypergraph Grammar with Graph Neural Network
Sep 28, 2023



Abstract:Property prediction plays an important role in material discovery. As an initial step to eventually develop a foundation model for material science, we introduce a new autoencoder called the MHG-GNN, which combines graph neural network (GNN) with Molecular Hypergraph Grammar (MHG). Results on a variety of property prediction tasks with diverse materials show that MHG-GNN is promising.
Biases in In Silico Evaluation of Molecular Optimization Methods and Bias-Reduced Evaluation Methodology
Jan 28, 2022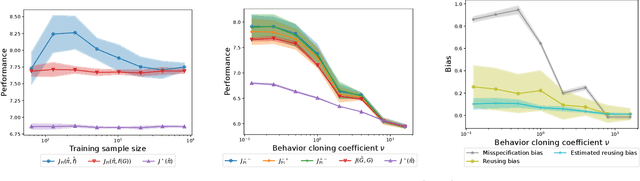
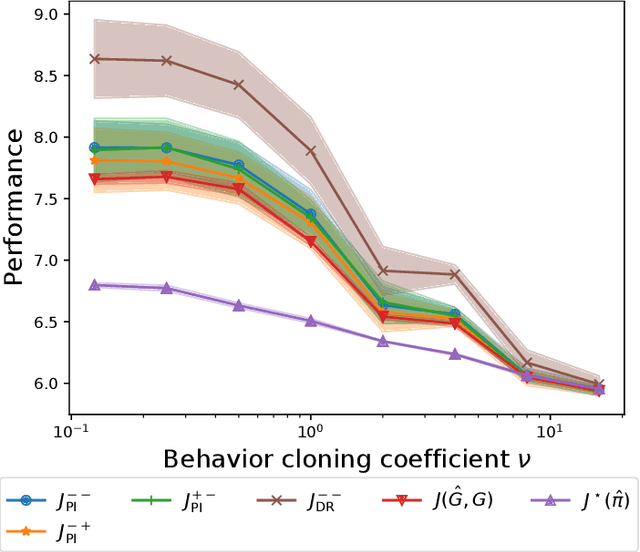
Abstract:We are interested in in silico evaluation methodology for molecular optimization methods. Given a sample of molecules and their properties of our interest, we wish not only to train an agent that can find molecules optimized with respect to the target property but also to evaluate its performance. A common practice is to train a predictor of the target property on the sample and use it for both training and evaluating the agent. We show that this evaluator potentially suffers from two biases; one is due to misspecification of the predictor and the other to reusing the same sample for training and evaluation. We discuss bias reduction methods for each of the biases comprehensively, and empirically investigate their effectiveness.
Classical Planning in Deep Latent Space
Jun 30, 2021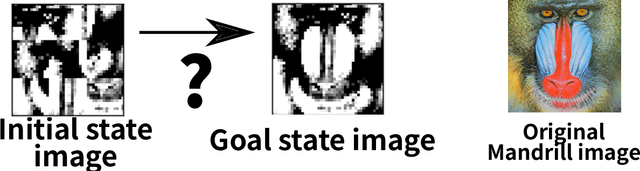
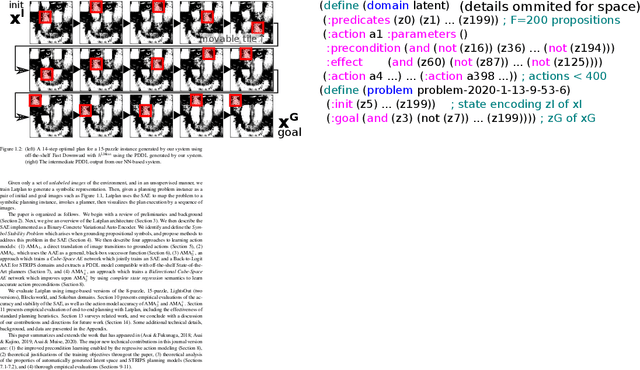
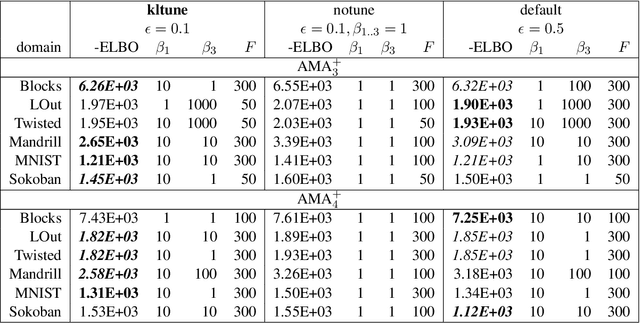
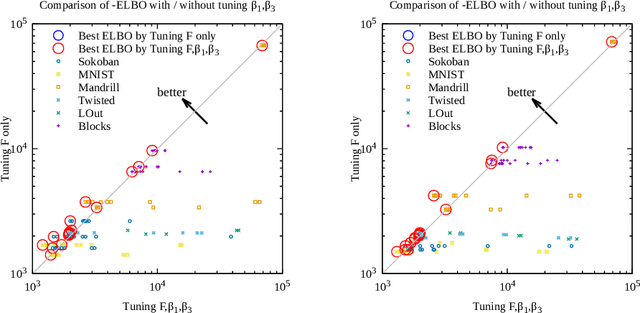
Abstract:Current domain-independent, classical planners require symbolic models of the problem domain and instance as input, resulting in a knowledge acquisition bottleneck. Meanwhile, although deep learning has achieved significant success in many fields, the knowledge is encoded in a subsymbolic representation which is incompatible with symbolic systems such as planners. We propose Latplan, an unsupervised architecture combining deep learning and classical planning. Given only an unlabeled set of image pairs showing a subset of transitions allowed in the environment (training inputs), Latplan learns a complete propositional PDDL action model of the environment. Later, when a pair of images representing the initial and the goal states (planning inputs) is given, Latplan finds a plan to the goal state in a symbolic latent space and returns a visualized plan execution. We evaluate Latplan using image-based versions of 6 planning domains: 8-puzzle, 15-Puzzle, Blocksworld, Sokoban and Two variations of LightsOut.
A Differentiable Point Process with Its Application to Spiking Neural Networks
Jun 03, 2021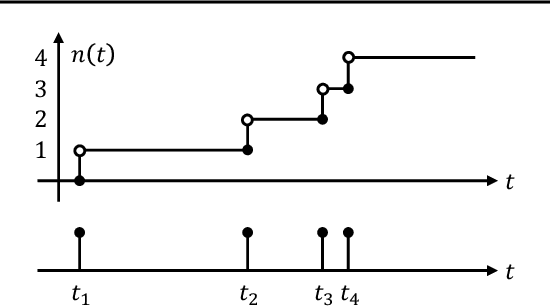
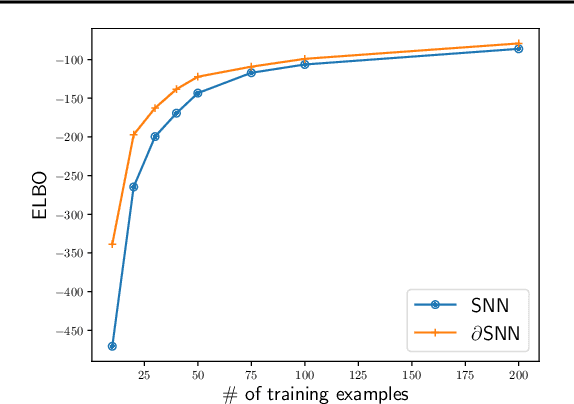
Abstract:This paper is concerned about a learning algorithm for a probabilistic model of spiking neural networks (SNNs). Jimenez Rezende & Gerstner (2014) proposed a stochastic variational inference algorithm to train SNNs with hidden neurons. The algorithm updates the variational distribution using the score function gradient estimator, whose high variance often impedes the whole learning algorithm. This paper presents an alternative gradient estimator for SNNs based on the path-wise gradient estimator. The main technical difficulty is a lack of a general method to differentiate a realization of an arbitrary point process, which is necessary to derive the path-wise gradient estimator. We develop a differentiable point process, which is the technical highlight of this paper, and apply it to derive the path-wise gradient estimator for SNNs. We investigate the effectiveness of our gradient estimator through numerical simulation.
Towards Stable Symbol Grounding with Zero-Suppressed State AutoEncoder
Mar 27, 2019



Abstract:While classical planning has been an active branch of AI, its applicability is limited to the tasks precisely modeled by humans. Fully automated high-level agents should be instead able to find a symbolic representation of an unknown environment without supervision, otherwise it exhibits the knowledge acquisition bottleneck. Meanwhile, Latplan (Asai and Fukunaga 2018) partially resolves the bottleneck with a neural network called State AutoEncoder (SAE). SAE obtains the propositional representation of the image-based puzzle domains with unsupervised learning, generates a state space and performs classical planning. In this paper, we identify the problematic, stochastic behavior of the SAE-produced propositions as a new sub-problem of symbol grounding problem, the symbol stability problem. Informally, symbols are stable when their referents (e.g. propositional values) do not change against small perturbation of the observation, and unstable symbols are harmful for symbolic reasoning. We analyze the problem in Latplan both formally and empirically, and propose "Zero-Suppressed SAE", an enhancement that stabilizes the propositions using the idea of closed-world assumption as a prior for NN optimization. We show that it finds the more stable propositions and the more compact representations, resulting in an improved success rate of Latplan. It is robust against various hyperparameters and eases the tuning effort, and also provides a weight pruning capability as a side effect.
Safe Exploration in Markov Decision Processes with Time-Variant Safety using Spatio-Temporal Gaussian Process
Sep 12, 2018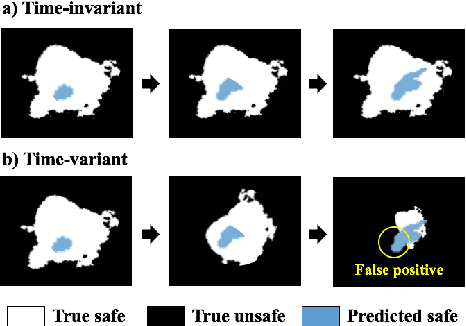

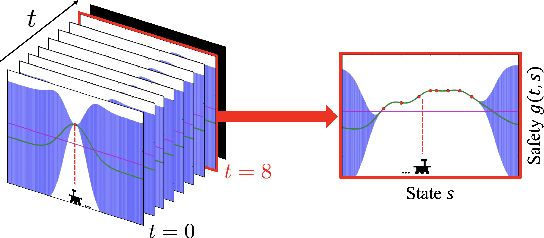

Abstract:In many real-world applications (e.g., planetary exploration, robot navigation), an autonomous agent must be able to explore a space with guaranteed safety. Most safe exploration algorithms in the field of reinforcement learning and robotics have been based on the assumption that the safety features are a priori known and time-invariant. This paper presents a learning algorithm called ST-SafeMDP for exploring Markov decision processes (MDPs) that is based on the assumption that the safety features are a priori unknown and time-variant. In this setting, the agent explores MDPs while constraining the probability of entering unsafe states defined by a safety function being below a threshold. The unknown and time-variant safety values are modeled using a spatio-temporal Gaussian process. However, there remains an issue that an agent may have no viable action in a shrinking true safe space. To address this issue, we formulate a problem maximizing the cumulative number of safe states in the worst case scenario with respect to future observations. The effectiveness of this approach was demonstrated in two simulation settings, including one using real lunar terrain data.
Molecular Hypergraph Grammar with its Application to Molecular Optimization
Sep 08, 2018



Abstract:This paper is concerned with a molecular optimization framework using variational autoencoders (VAEs). In this paradigm, VAE allows us to convert a molecular graph into/from its latent continuous vector, and therefore, the molecular optimization problem can be solved by continuous optimization techniques. One of the longstanding issues in this area is that it is difficult to always generate valid molecules. The very recent work called the junction tree variational autoencoder (JT-VAE) successfully solved this issue by generating a molecule fragment-by-fragment. While it achieves the state-of-the-art performance, it requires several neural networks to be trained, which predict which atoms are used to connect fragments and stereochemistry of each bond. In this paper, we present a molecular hypergraph grammar variational autoencoder (MHG-VAE), which uses a single VAE to address the issue. Our idea is to develop a novel graph grammar for molecular graphs called molecular hypergraph grammar (MHG), which can specify the connections between fragments and the stereochemistry on behalf of neural networks. This capability allows us to address the issue using only a single VAE. We empirically demonstrate the effectiveness of MHG-VAE over existing methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge