Hassan Rivaz
Singular Value Few-shot Adaptation of Vision-Language Models
Sep 03, 2025Abstract:Vision-language models (VLMs) like CLIP have shown impressive zero-shot and few-shot learning capabilities across diverse applications. However, adapting these models to new fine-grained domains remains difficult due to reliance on prompt engineering and the high cost of full model fine-tuning. Existing adaptation approaches rely on augmented components, such as prompt tokens and adapter modules, which could limit adaptation quality, destabilize the model, and compromise the rich knowledge learned during pretraining. In this work, we present \textbf{CLIP-SVD}, a novel \textit{multi-modal} and \textit{parameter-efficient} adaptation technique that leverages Singular Value Decomposition (SVD) to modify the internal parameter space of CLIP without injecting additional modules. Specifically, we fine-tune only the singular values of the CLIP parameter matrices to rescale the basis vectors for domain adaptation while retaining the pretrained model. This design enables enhanced adaptation performance using only \textbf{0.04\%} of the model's total parameters and better preservation of its generalization ability. CLIP-SVD achieves state-of-the-art classification results on 11 natural and 10 biomedical datasets, outperforming previous methods in both accuracy and generalization under few-shot settings. Additionally, we leverage a natural language-based approach to analyze the effectiveness and dynamics of the CLIP adaptation to allow interpretability of CLIP-SVD. The code is publicly available at https://github.com/HealthX-Lab/CLIP-SVD.
DINOMotion: advanced robust tissue motion tracking with DINOv2 in 2D-Cine MRI-guided radiotherapy
Aug 14, 2025Abstract:Accurate tissue motion tracking is critical to ensure treatment outcome and safety in 2D-Cine MRI-guided radiotherapy. This is typically achieved by registration of sequential images, but existing methods often face challenges with large misalignments and lack of interpretability. In this paper, we introduce DINOMotion, a novel deep learning framework based on DINOv2 with Low-Rank Adaptation (LoRA) layers for robust, efficient, and interpretable motion tracking. DINOMotion automatically detects corresponding landmarks to derive optimal image registration, enhancing interpretability by providing explicit visual correspondences between sequential images. The integration of LoRA layers reduces trainable parameters, improving training efficiency, while DINOv2's powerful feature representations offer robustness against large misalignments. Unlike iterative optimization-based methods, DINOMotion directly computes image registration at test time. Our experiments on volunteer and patient datasets demonstrate its effectiveness in estimating both linear and nonlinear transformations, achieving Dice scores of 92.07% for the kidney, 90.90% for the liver, and 95.23% for the lung, with corresponding Hausdorff distances of 5.47 mm, 8.31 mm, and 6.72 mm, respectively. DINOMotion processes each scan in approximately 30ms and consistently outperforms state-of-the-art methods, particularly in handling large misalignments. These results highlight its potential as a robust and interpretable solution for real-time motion tracking in 2D-Cine MRI-guided radiotherapy.
Weakly Supervised Intracranial Aneurysm Detection and Segmentation in MR angiography via Multi-task UNet with Vesselness Prior
Aug 01, 2025Abstract:Intracranial aneurysms (IAs) are abnormal dilations of cerebral blood vessels that, if ruptured, can lead to life-threatening consequences. However, their small size and soft contrast in radiological scans often make it difficult to perform accurate and efficient detection and morphological analyses, which are critical in the clinical care of the disorder. Furthermore, the lack of large public datasets with voxel-wise expert annotations pose challenges for developing deep learning algorithms to address the issues. Therefore, we proposed a novel weakly supervised 3D multi-task UNet that integrates vesselness priors to jointly perform aneurysm detection and segmentation in time-of-flight MR angiography (TOF-MRA). Specifically, to robustly guide IA detection and segmentation, we employ the popular Frangi's vesselness filter to derive soft cerebrovascular priors for both network input and an attention block to conduct segmentation from the decoder and detection from an auxiliary branch. We train our model on the Lausanne dataset with coarse ground truth segmentation, and evaluate it on the test set with refined labels from the same database. To further assess our model's generalizability, we also validate it externally on the ADAM dataset. Our results demonstrate the superior performance of the proposed technique over the SOTA techniques for aneurysm segmentation (Dice = 0.614, 95%HD =1.38mm) and detection (false positive rate = 1.47, sensitivity = 92.9%).
TextSAM-EUS: Text Prompt Learning for SAM to Accurately Segment Pancreatic Tumor in Endoscopic Ultrasound
Jul 24, 2025Abstract:Pancreatic cancer carries a poor prognosis and relies on endoscopic ultrasound (EUS) for targeted biopsy and radiotherapy. However, the speckle noise, low contrast, and unintuitive appearance of EUS make segmentation of pancreatic tumors with fully supervised deep learning (DL) models both error-prone and dependent on large, expert-curated annotation datasets. To address these challenges, we present TextSAM-EUS, a novel, lightweight, text-driven adaptation of the Segment Anything Model (SAM) that requires no manual geometric prompts at inference. Our approach leverages text prompt learning (context optimization) through the BiomedCLIP text encoder in conjunction with a LoRA-based adaptation of SAM's architecture to enable automatic pancreatic tumor segmentation in EUS, tuning only 0.86% of the total parameters. On the public Endoscopic Ultrasound Database of the Pancreas, TextSAM-EUS with automatic prompts attains 82.69% Dice and 85.28% normalized surface distance (NSD), and with manual geometric prompts reaches 83.10% Dice and 85.70% NSD, outperforming both existing state-of-the-art (SOTA) supervised DL models and foundation models (e.g., SAM and its variants). As the first attempt to incorporate prompt learning in SAM-based medical image segmentation, TextSAM-EUS offers a practical option for efficient and robust automatic EUS segmentation. Our code will be publicly available upon acceptance.
Lightweight Physics-Informed Zero-Shot Ultrasound Plane Wave Denoising
Jun 26, 2025Abstract:Ultrasound Coherent Plane Wave Compounding (CPWC) enhances image contrast by combining echoes from multiple steered transmissions. While increasing the number of angles generally improves image quality, it drastically reduces the frame rate and can introduce blurring artifacts in fast-moving targets. Moreover, compounded images remain susceptible to noise, particularly when acquired with a limited number of transmissions. We propose a zero-shot denoising framework tailored for low-angle CPWC acquisitions, which enhances contrast without relying on a separate training dataset. The method divides the available transmission angles into two disjoint subsets, each used to form compound images that include higher noise levels. The new compounded images are then used to train a deep model via a self-supervised residual learning scheme, enabling it to suppress incoherent noise while preserving anatomical structures. Because angle-dependent artifacts vary between the subsets while the underlying tissue response is similar, this physics-informed pairing allows the network to learn to disentangle the inconsistent artifacts from the consistent tissue signal. Unlike supervised methods, our model requires no domain-specific fine-tuning or paired data, making it adaptable across anatomical regions and acquisition setups. The entire pipeline supports efficient training with low computational cost due to the use of a lightweight architecture, which comprises only two convolutional layers. Evaluations on simulation, phantom, and in vivo data demonstrate superior contrast enhancement and structure preservation compared to both classical and deep learning-based denoising methods.
Medical Image Classification with KAN-Integrated Transformers and Dilated Neighborhood Attention
Feb 19, 2025Abstract:Convolutional networks, transformers, hybrid models, and Mamba-based architectures have demonstrated strong performance across various medical image classification tasks. However, these methods were primarily designed to classify clean images using labeled data. In contrast, real-world clinical data often involve image corruptions that are unique to multi-center studies and stem from variations in imaging equipment across manufacturers. In this paper, we introduce the Medical Vision Transformer (MedViTV2), a novel architecture incorporating Kolmogorov-Arnold Network (KAN) layers into the transformer architecture for the first time, aiming for generalized medical image classification. We have developed an efficient KAN block to reduce computational load while enhancing the accuracy of the original MedViT. Additionally, to counteract the fragility of our MedViT when scaled up, we propose an enhanced Dilated Neighborhood Attention (DiNA), an adaptation of the efficient fused dot-product attention kernel capable of capturing global context and expanding receptive fields to scale the model effectively and addressing feature collapse issues. Moreover, a hierarchical hybrid strategy is introduced to stack our Local Feature Perception and Global Feature Perception blocks in an efficient manner, which balances local and global feature perceptions to boost performance. Extensive experiments on 17 medical image classification datasets and 12 corrupted medical image datasets demonstrate that MedViTV2 achieved state-of-the-art results in 27 out of 29 experiments with reduced computational complexity. MedViTV2 is 44\% more computationally efficient than the previous version and significantly enhances accuracy, achieving improvements of 4.6\% on MedMNIST, 5.8\% on NonMNIST, and 13.4\% on the MedMNIST-C benchmark.
Ultrasound Image Generation using Latent Diffusion Models
Feb 12, 2025Abstract:Diffusion models for image generation have been a subject of increasing interest due to their ability to generate diverse, high-quality images. Image generation has immense potential in medical imaging because open-source medical images are difficult to obtain compared to natural images, especially for rare conditions. The generated images can be used later to train classification and segmentation models. In this paper, we propose simulating realistic ultrasound (US) images by successive fine-tuning of large diffusion models on different publicly available databases. To do so, we fine-tuned Stable Diffusion, a state-of-the-art latent diffusion model, on BUSI (Breast US Images) an ultrasound breast image dataset. We successfully generated high-quality US images of the breast using simple prompts that specify the organ and pathology, which appeared realistic to three experienced US scientists and a US radiologist. Additionally, we provided user control by conditioning the model with segmentations through ControlNet. We will release the source code at http://code.sonography.ai/ to allow fast US image generation to the scientific community.
Constrained and Regularized Quantitative Ultrasound Parameter Estimation using ADMM
Jan 07, 2025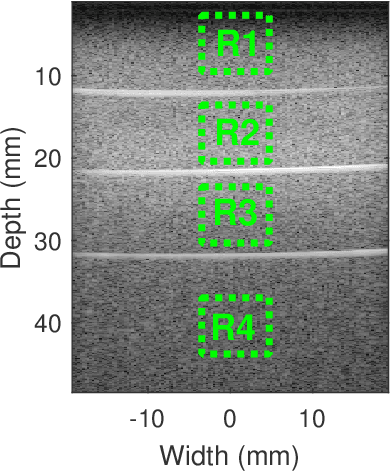

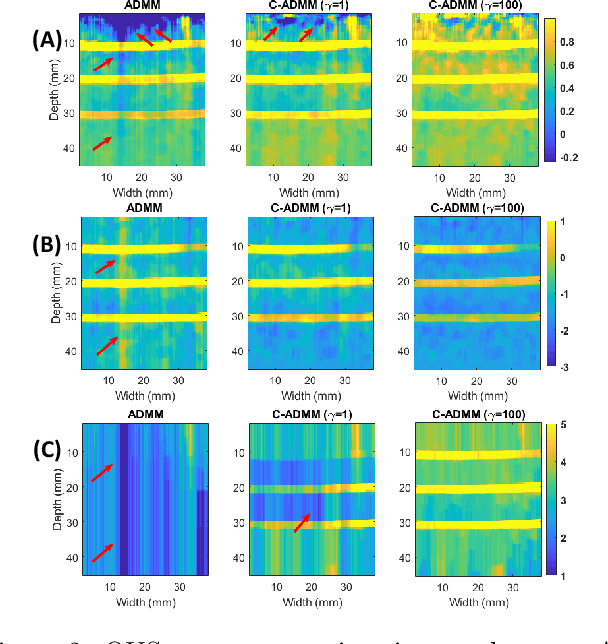

Abstract:Regularized estimation of quantitative ultrasound (QUS) parameters, such as attenuation and backscatter coefficients, has gained research interest. Recently, the alternating direction method of multipliers (ADMM) has been applied successfully to estimate these parameters, by utilizing L2 and L1 norms for attenuation and backscatter coefficient regularization, respectively. While this method improves upon previous approaches, it does not fully leverage the prior knowledge of minimum physically feasible parameter values, sometimes yielding values outside the realistic range. This work addresses this limitation by incorporating minimum QUS parameter values as constraints to enhance ADMM estimation. The proposed method is validated using experimental phantom data.
Reliability of deep learning models for anatomical landmark detection: The role of inter-rater variability
Nov 26, 2024
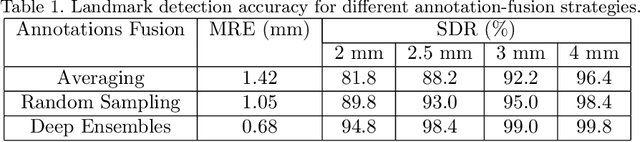
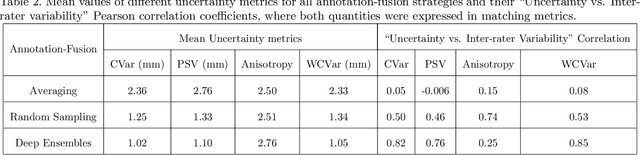
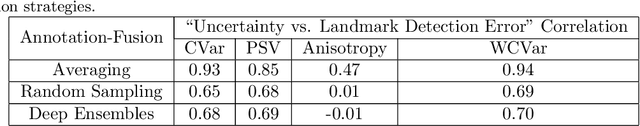
Abstract:Automated detection of anatomical landmarks plays a crucial role in many diagnostic and surgical applications. Progresses in deep learning (DL) methods have resulted in significant performance enhancement in tasks related to anatomical landmark detection. While current research focuses on accurately localizing these landmarks in medical scans, the importance of inter-rater annotation variability in building DL models is often overlooked. Understanding how inter-rater variability impacts the performance and reliability of the resulting DL algorithms, which are crucial for clinical deployment, can inform the improvement of training data construction and boost DL models' outcomes. In this paper, we conducted a thorough study of different annotation-fusion strategies to preserve inter-rater variability in DL models for anatomical landmark detection, aiming to boost the performance and reliability of the resulting algorithms. Additionally, we explored the characteristics and reliability of four metrics, including a novel Weighted Coordinate Variance metric to quantify landmark detection uncertainty/inter-rater variability. Our research highlights the crucial connection between inter-rater variability, DL-models performances, and uncertainty, revealing how different approaches for multi-rater landmark annotation fusion can influence these factors.
CAMLD: Contrast-Agnostic Medical Landmark Detection with Consistency-Based Regularization
Nov 26, 2024Abstract:Anatomical landmark detection in medical images is essential for various clinical and research applications, including disease diagnosis and surgical planning. However, manual landmark annotation is time-consuming and requires significant expertise. Existing deep learning (DL) methods often require large amounts of well-annotated data, which are costly to acquire. In this paper, we introduce CAMLD, a novel self-supervised DL framework for anatomical landmark detection in unlabeled scans with varying contrasts by using only a single reference example. To achieve this, we employed an inter-subject landmark consistency loss with an image registration loss while introducing a 3D convolution-based contrast augmentation strategy to promote model generalization to new contrasts. Additionally, we utilize an adaptive mixed loss function to schedule the contributions of different sub-tasks for optimal outcomes. We demonstrate the proposed method with the intricate task of MRI-based 3D brain landmark detection. With comprehensive experiments on four diverse clinical and public datasets, including both T1w and T2w MRI scans at different MRI field strengths, we demonstrate that CAMLD outperforms the state-of-the-art methods in terms of mean radial errors (MREs) and success detection rates (SDRs). Our framework provides a robust and accurate solution for anatomical landmark detection, reducing the need for extensively annotated datasets and generalizing well across different imaging contrasts. Our code will be publicly available at: https://github.com/HealthX-Lab/CAMLD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge