Mostafa Sharifzadeh
Lightweight Physics-Informed Zero-Shot Ultrasound Plane Wave Denoising
Jun 26, 2025Abstract:Ultrasound Coherent Plane Wave Compounding (CPWC) enhances image contrast by combining echoes from multiple steered transmissions. While increasing the number of angles generally improves image quality, it drastically reduces the frame rate and can introduce blurring artifacts in fast-moving targets. Moreover, compounded images remain susceptible to noise, particularly when acquired with a limited number of transmissions. We propose a zero-shot denoising framework tailored for low-angle CPWC acquisitions, which enhances contrast without relying on a separate training dataset. The method divides the available transmission angles into two disjoint subsets, each used to form compound images that include higher noise levels. The new compounded images are then used to train a deep model via a self-supervised residual learning scheme, enabling it to suppress incoherent noise while preserving anatomical structures. Because angle-dependent artifacts vary between the subsets while the underlying tissue response is similar, this physics-informed pairing allows the network to learn to disentangle the inconsistent artifacts from the consistent tissue signal. Unlike supervised methods, our model requires no domain-specific fine-tuning or paired data, making it adaptable across anatomical regions and acquisition setups. The entire pipeline supports efficient training with low computational cost due to the use of a lightweight architecture, which comprises only two convolutional layers. Evaluations on simulation, phantom, and in vivo data demonstrate superior contrast enhancement and structure preservation compared to both classical and deep learning-based denoising methods.
Denoising Plane Wave Ultrasound Images Using Diffusion Probabilistic Models
Aug 20, 2024



Abstract:Ultrasound plane wave imaging is a cutting-edge technique that enables high frame-rate imaging. However, one challenge associated with high frame-rate ultrasound imaging is the high noise associated with them, hindering their wider adoption. Therefore, the development of a denoising method becomes imperative to augment the quality of plane wave images. Drawing inspiration from Denoising Diffusion Probabilistic Models (DDPMs), our proposed solution aims to enhance plane wave image quality. Specifically, the method considers the distinction between low-angle and high-angle compounding plane waves as noise and effectively eliminates it by adapting a DDPM to beamformed radiofrequency (RF) data. The method underwent training using only 400 simulated images. In addition, our approach employs natural image segmentation masks as intensity maps for the generated images, resulting in accurate denoising for various anatomy shapes. The proposed method was assessed across simulation, phantom, and in vivo images. The results of the evaluations indicate that our approach not only enhances image quality on simulated data but also demonstrates effectiveness on phantom and in vivo data in terms of image quality. Comparative analysis with other methods underscores the superiority of our proposed method across various evaluation metrics. The source code and trained model will be released along with the dataset at: http://code.sonography.ai
Robust RF Data Normalization for Deep Learning
Aug 22, 2023



Abstract:Radio frequency (RF) data contain richer information compared to other data types, such as envelope or B-mode, and employing RF data for training deep neural networks has attracted growing interest in ultrasound image processing. However, RF data is highly fluctuating and additionally has a high dynamic range. Most previous studies in the literature have relied on conventional data normalization, which has been adopted within the computer vision community. We demonstrate the inadequacy of those techniques for normalizing RF data and propose that individual standardization of each image substantially enhances the performance of deep neural networks by utilizing the data more efficiently. We compare conventional and proposed normalizations in a phase aberration correction task and illustrate how the former enhances the generality of trained models.
Frequency-Space Prediction Filtering for Phase Aberration Correction in Plane-Wave Ultrasound
Aug 22, 2023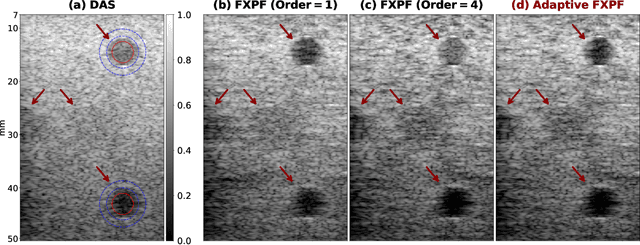
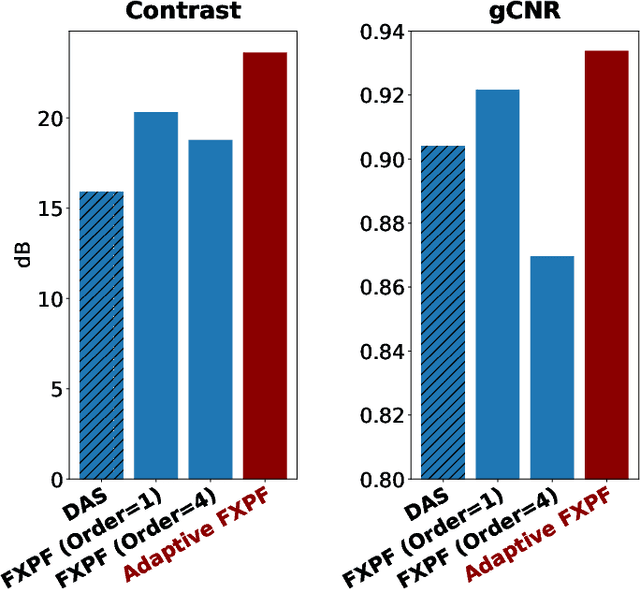
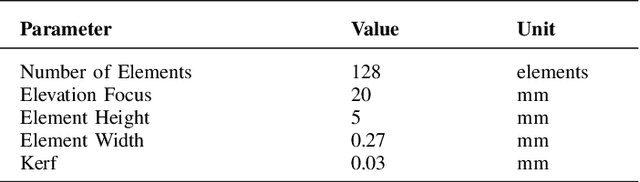
Abstract:Ultrasound imaging often suffers from image degradation stemming from phase aberration, which represents a significant contributing factor to the overall image degradation in ultrasound imaging. Frequency-space prediction filtering or FXPF is a technique that has been applied within focused ultrasound imaging to alleviate the phase aberration effect. It presupposes the existence of an autoregressive (AR) model across the signals received at the transducer elements and removes any components that do not conform to the established model. In this study, we illustrate the challenge of applying this technique to plane-wave imaging, where, at shallower depths, signals from more distant elements lose relevance, and a fewer number of elements contribute to image reconstruction. While the number of contributing signals varies, adopting a fixed-order AR model across all depths, results in suboptimal performance. To address this challenge, we propose an AR model with an adaptive order and quantify its effectiveness using contrast and generalized contrast-to-noise ratio metrics.
Phase Aberration Correction: A Deep Learning-Based Aberration to Aberration Approach
Aug 22, 2023



Abstract:One of the primary sources of suboptimal image quality in ultrasound imaging is phase aberration. It is caused by spatial changes in sound speed over a heterogeneous medium, which disturbs the transmitted waves and prevents coherent summation of echo signals. Obtaining non-aberrated ground truths in real-world scenarios can be extremely challenging, if not impossible. This challenge hinders training of deep learning-based techniques' performance due to the presence of domain shift between simulated and experimental data. Here, for the first time, we propose a deep learning-based method that does not require ground truth to correct the phase aberration problem, and as such, can be directly trained on real data. We train a network wherein both the input and target output are randomly aberrated radio frequency (RF) data. Moreover, we demonstrate that a conventional loss function such as mean square error is inadequate for training such a network to achieve optimal performance. Instead, we propose an adaptive mixed loss function that employs both B-mode and RF data, resulting in more efficient convergence and enhanced performance. Finally, we publicly release our dataset, including 161,701 single plane-wave images (RF data). This dataset serves to mitigate the data scarcity problem in the development of deep learning-based techniques for phase aberration correction.
Phase Aberration Correction without Reference Data: An Adaptive Mixed Loss Deep Learning Approach
Mar 10, 2023Abstract:Phase aberration is one of the primary sources of image quality degradation in ultrasound, which is induced by spatial variations in sound speed across the heterogeneous medium. This effect disrupts transmitted waves and prevents coherent summation of echo signals, resulting in suboptimal image quality. In real experiments, obtaining non-aberrated ground truths can be extremely challenging, if not infeasible. It hinders the performance of deep learning-based phase aberration correction techniques due to sole reliance on simulated data and the presence of domain shift between simulated and experimental data. Here, for the first time, we propose a deep learning-based method that does not require reference data to compensate for the phase aberration effect. We train a network wherein both input and target output are randomly aberrated radio frequency (RF) data. Moreover, we demonstrate that a conventional loss function such as mean square error is inadequate for training the network to achieve optimal performance. Instead, we propose an adaptive mixed loss function that employs both B-mode and RF data, resulting in more efficient convergence and enhanced performance. Source code is available at \url{http://code.sonography.ai}.
Bi-Directional Semi-Supervised Training of Convolutional Neural Networks for Ultrasound Elastography Displacement Estimation
Jan 31, 2022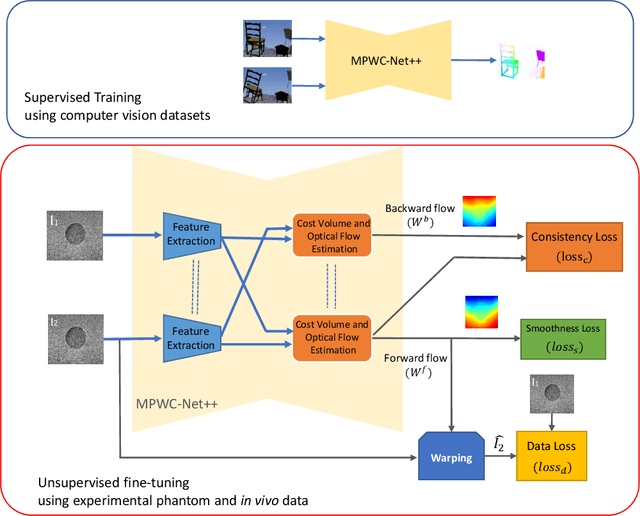
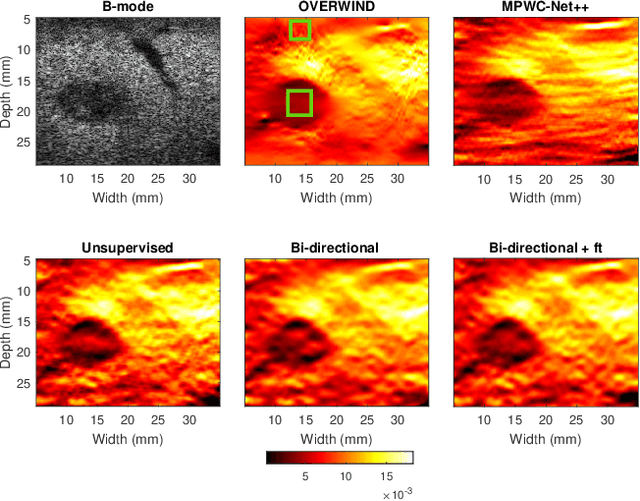
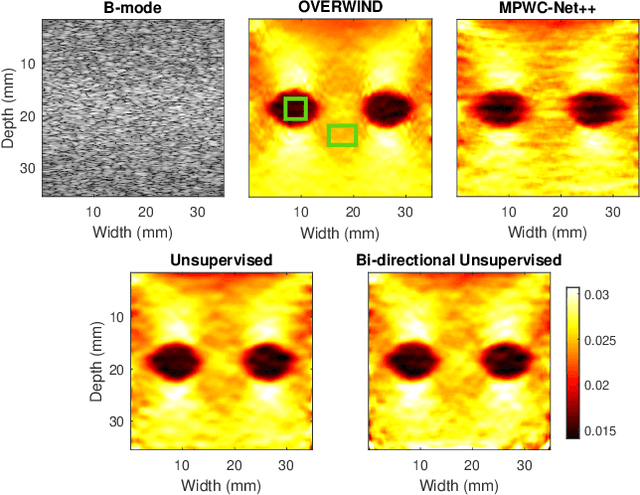
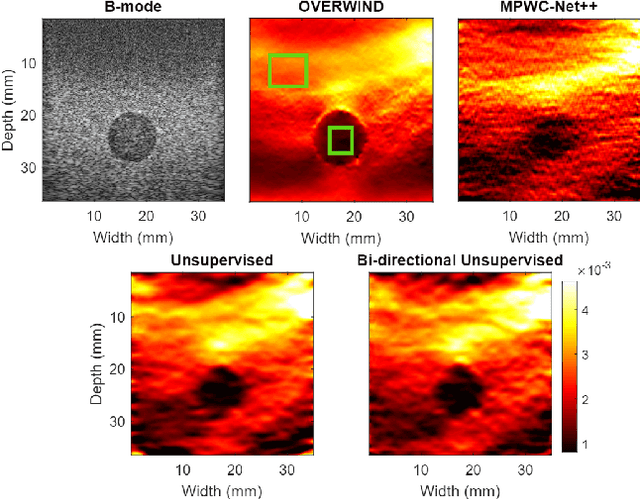
Abstract:The performance of ultrasound elastography (USE) heavily depends on the accuracy of displacement estimation. Recently, Convolutional Neural Networks (CNN) have shown promising performance in optical flow estimation and have been adopted for USE displacement estimation. Networks trained on computer vision images are not optimized for USE displacement estimation since there is a large gap between the computer vision images and the high-frequency Radio Frequency (RF) ultrasound data. Many researchers tried to adopt the optical flow CNNs to USE by applying transfer learning to improve the performance of CNNs for USE. However, the ground truth displacement in real ultrasound data is unknown, and simulated data exhibits a domain shift compared to the real data and is also computationally expensive to generate. To resolve this issue, semi-supervised methods have been proposed wherein the networks pre-trained on computer vision images are fine-tuned using real ultrasound data. In this paper, we employ a semi-supervised method by exploiting the first and second-order derivatives of the displacement field for the regularization. We also modify the network structure to estimate both forward and backward displacements, and propose to use consistency between the forward and backward strains as an additional regularizer to further enhance the performance. We validate our method using several experimental phantom and in vivo data. We also show that the network fine-tuned by our proposed method using experimental phantom data performs well on in vivo data similar to the network fine-tuned on in vivo data. Our results also show that the proposed method outperforms current deep learning methods and is comparable to computationally expensive optimization-based algorithms.
Ultrasound Domain Adaptation Using Frequency Domain Analysis
Sep 21, 2021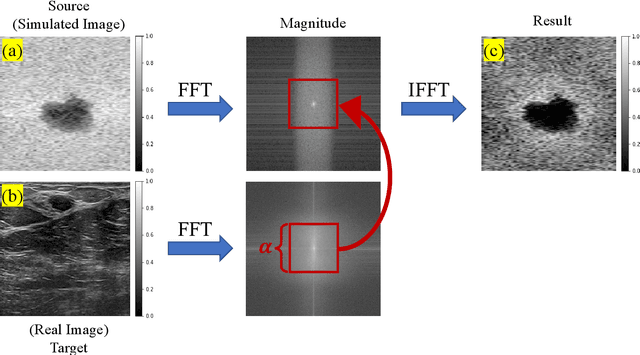
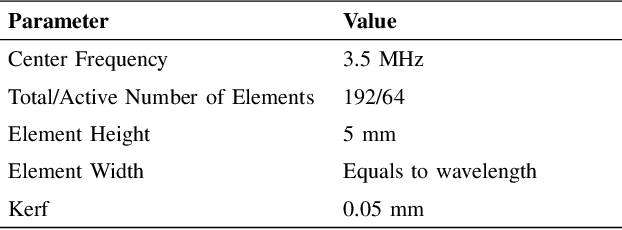
Abstract:A common issue in exploiting simulated ultrasound data for training neural networks is the domain shift problem, where the trained models on synthetic data are not generalizable to clinical data. Recently, Fourier Domain Adaptation (FDA) has been proposed in the field of computer vision to tackle the domain shift problem by replacing the magnitude of the low-frequency spectrum of a synthetic sample (source) with a real sample (target). This method is attractive in ultrasound imaging given that two important differences between synthetic and real ultrasound data are caused by unknown values of attenuation and speed of sound (SOS) in real tissues. Attenuation leads to slow variations in the amplitude of the B-mode image, and SOS mismatch creates aberration and subsequent blurring. As such, both domain shifts cause differences in the low-frequency components of the envelope data, which are replaced in the proposed method. We demonstrate that applying the FDA method to the synthetic data, simulated by Field II, obtains an 3.5\% higher Dice similarity coefficient for a breast lesion segmentation task.
An Ultra-Fast Method for Simulation of Realistic Ultrasound Images
Sep 21, 2021

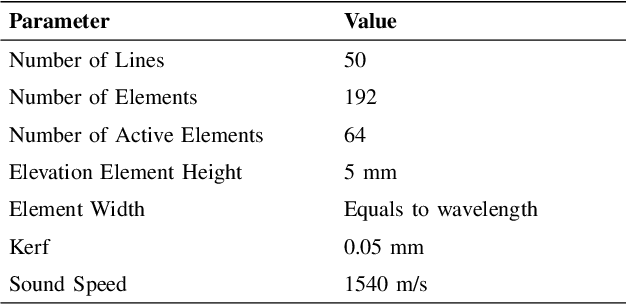
Abstract:Convolutional neural networks (CNNs) have attracted a rapidly growing interest in a variety of different processing tasks in the medical ultrasound community. However, the performance of CNNs is highly reliant on both the amount and fidelity of the training data. Therefore, scarce data is almost always a concern, particularly in the medical field, where clinical data is not easily accessible. The utilization of synthetic data is a popular approach to address this challenge. However, but simulating a large number of images using packages such as Field II is time-consuming, and the distribution of simulated images is far from that of the real images. Herein, we introduce a novel ultra-fast ultrasound image simulation method based on the Fourier transform and evaluate its performance in a lesion segmentation task. We demonstrate that data augmentation using the images generated by the proposed method substantially outperforms Field II in terms of Dice similarity coefficient, while the simulation is almost 36000 times faster (both on CPU).
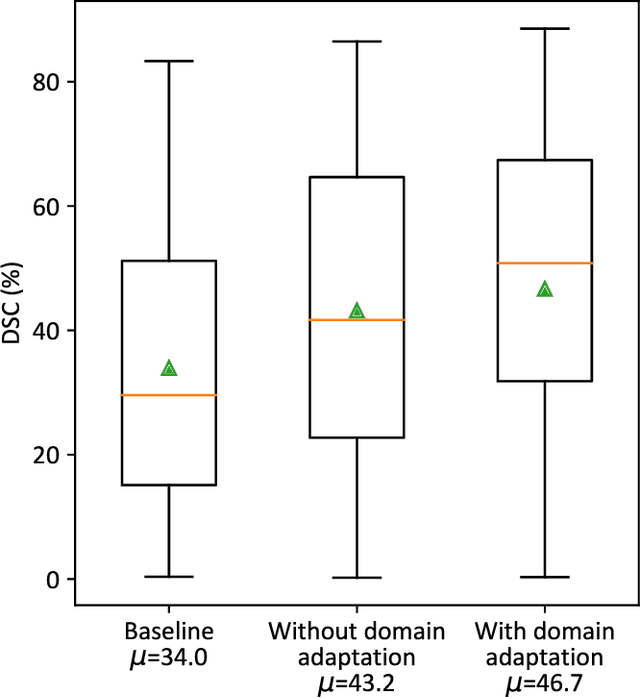
Investigating Shift-Variance of Convolutional Neural Networks in Ultrasound Image Segmentation
Jul 22, 2021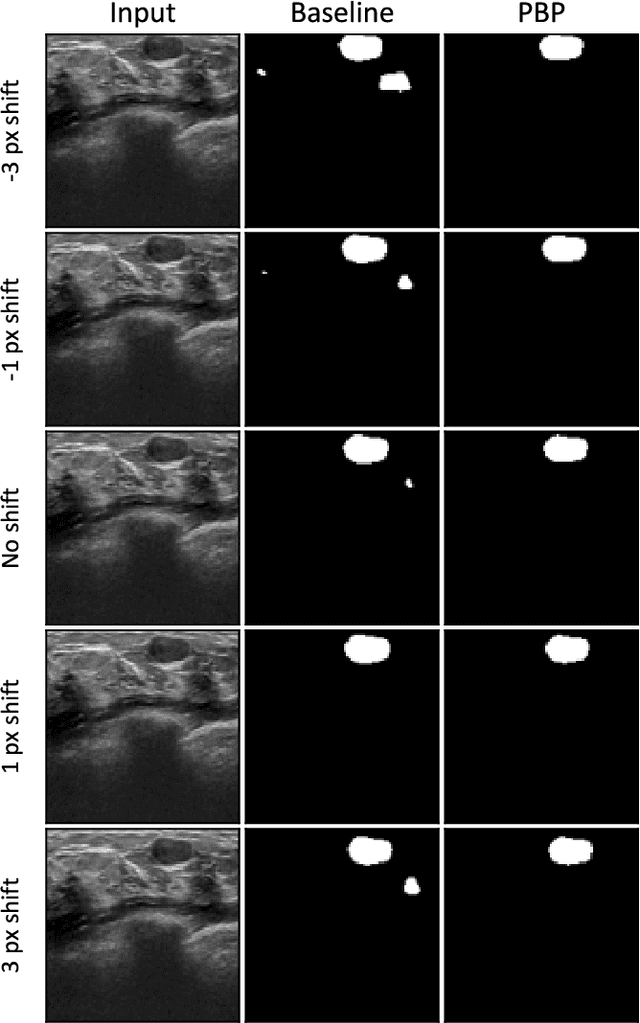
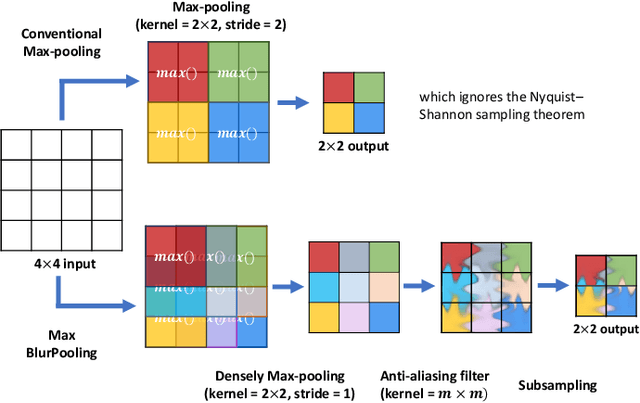
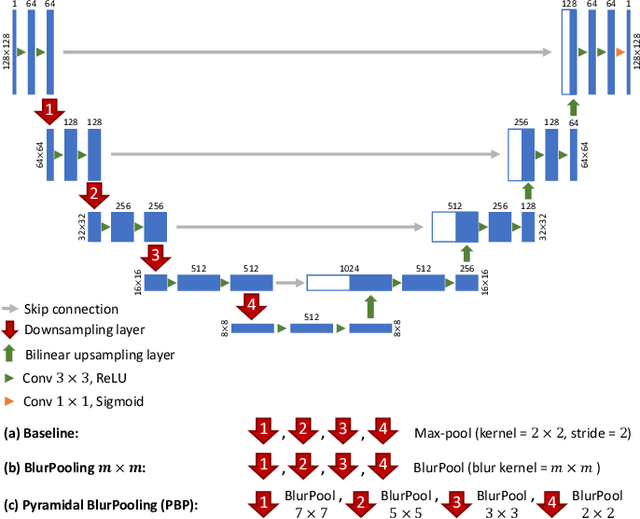
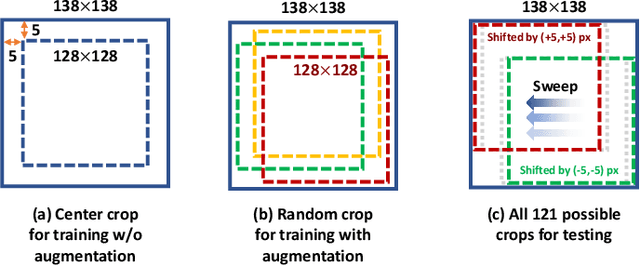
Abstract:While accuracy is an evident criterion for ultrasound image segmentation, output consistency across different tests is equally crucial for tracking changes in regions of interest in applications such as monitoring the patients' response to treatment, measuring the progression or regression of the disease, reaching a diagnosis, or treatment planning. Convolutional neural networks (CNNs) have attracted rapidly growing interest in automatic ultrasound image segmentation recently. However, CNNs are not shift-equivariant, meaning that if the input translates, e.g., in the lateral direction by one pixel, the output segmentation may drastically change. To the best of our knowledge, this problem has not been studied in ultrasound image segmentation or even more broadly in ultrasound images. Herein, we investigate and quantify the shift-variance problem of CNNs in this application and further evaluate the performance of a recently published technique, called BlurPooling, for addressing the problem. In addition, we propose the Pyramidal BlurPooling method that outperforms BlurPooling in both output consistency and segmentation accuracy. Finally, we demonstrate that data augmentation is not a replacement for the proposed method. Source code is available at https://git.io/pbpunet and http://code.sonography.ai.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge