Haihong Yang
LLM-Mini-CEX: Automatic Evaluation of Large Language Model for Diagnostic Conversation
Aug 15, 2023Abstract:There is an increasing interest in developing LLMs for medical diagnosis to improve diagnosis efficiency. Despite their alluring technological potential, there is no unified and comprehensive evaluation criterion, leading to the inability to evaluate the quality and potential risks of medical LLMs, further hindering the application of LLMs in medical treatment scenarios. Besides, current evaluations heavily rely on labor-intensive interactions with LLMs to obtain diagnostic dialogues and human evaluation on the quality of diagnosis dialogue. To tackle the lack of unified and comprehensive evaluation criterion, we first initially establish an evaluation criterion, termed LLM-specific Mini-CEX to assess the diagnostic capabilities of LLMs effectively, based on original Mini-CEX. To address the labor-intensive interaction problem, we develop a patient simulator to engage in automatic conversations with LLMs, and utilize ChatGPT for evaluating diagnosis dialogues automatically. Experimental results show that the LLM-specific Mini-CEX is adequate and necessary to evaluate medical diagnosis dialogue. Besides, ChatGPT can replace manual evaluation on the metrics of humanistic qualities and provides reproducible and automated comparisons between different LLMs.
Molecular Contrastive Learning with Chemical Element Knowledge Graph
Dec 01, 2021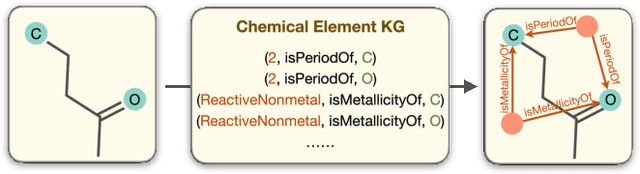
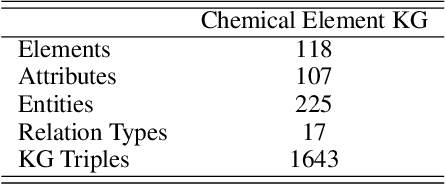
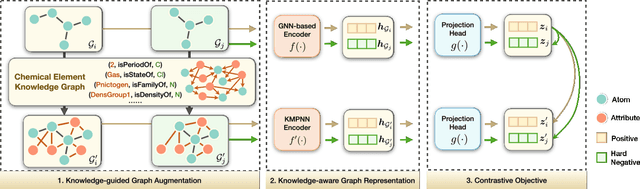
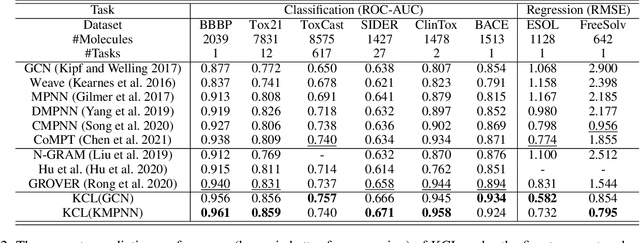
Abstract:Molecular representation learning contributes to multiple downstream tasks such as molecular property prediction and drug design. To properly represent molecules, graph contrastive learning is a promising paradigm as it utilizes self-supervision signals and has no requirements for human annotations. However, prior works fail to incorporate fundamental domain knowledge into graph semantics and thus ignore the correlations between atoms that have common attributes but are not directly connected by bonds. To address these issues, we construct a Chemical Element Knowledge Graph (KG) to summarize microscopic associations between elements and propose a novel Knowledge-enhanced Contrastive Learning (KCL) framework for molecular representation learning. KCL framework consists of three modules. The first module, knowledge-guided graph augmentation, augments the original molecular graph based on the Chemical Element KG. The second module, knowledge-aware graph representation, extracts molecular representations with a common graph encoder for the original molecular graph and a Knowledge-aware Message Passing Neural Network (KMPNN) to encode complex information in the augmented molecular graph. The final module is a contrastive objective, where we maximize agreement between these two views of molecular graphs. Extensive experiments demonstrated that KCL obtained superior performances against state-of-the-art baselines on eight molecular datasets. Visualization experiments properly interpret what KCL has learned from atoms and attributes in the augmented molecular graphs. Our codes and data are available in supplementary materials.
Knowledge-aware Contrastive Molecular Graph Learning
Mar 24, 2021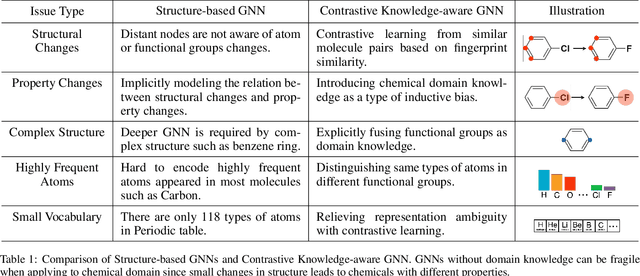
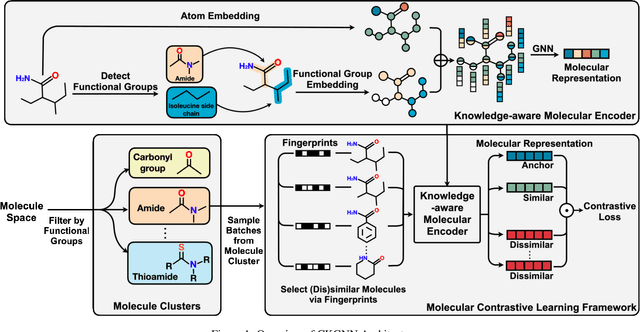
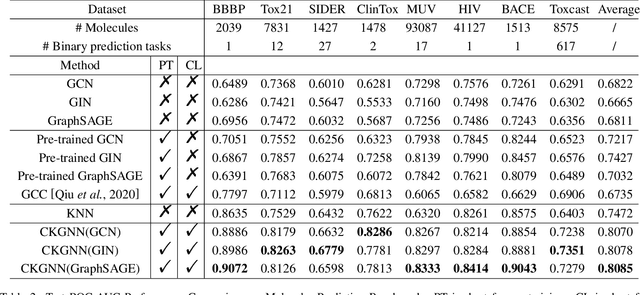
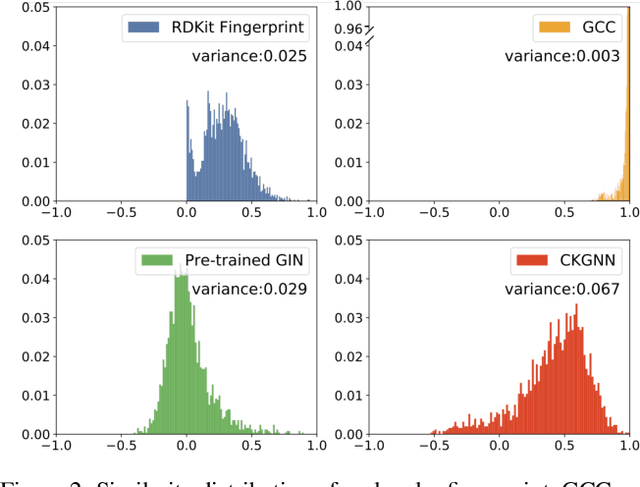
Abstract:Leveraging domain knowledge including fingerprints and functional groups in molecular representation learning is crucial for chemical property prediction and drug discovery. When modeling the relation between graph structure and molecular properties implicitly, existing works can hardly capture structural or property changes and complex structure, with much smaller atom vocabulary and highly frequent atoms. In this paper, we propose the Contrastive Knowledge-aware GNN (CKGNN) for self-supervised molecular representation learning to fuse domain knowledge into molecular graph representation. We explicitly encode domain knowledge via knowledge-aware molecular encoder under the contrastive learning framework, ensuring that the generated molecular embeddings equipped with chemical domain knowledge to distinguish molecules with similar chemical formula but dissimilar functions. Extensive experiments on 8 public datasets demonstrate the effectiveness of our model with a 6\% absolute improvement on average against strong competitors. Ablation study and further investigation also verify the best of both worlds: incorporation of chemical domain knowledge into self-supervised learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge