Gregory Darnell
RelCon: Relative Contrastive Learning for a Motion Foundation Model for Wearable Data
Nov 27, 2024



Abstract:We present RelCon, a novel self-supervised \textit{Rel}ative \textit{Con}trastive learning approach that uses a learnable distance measure in combination with a softened contrastive loss for training an motion foundation model from wearable sensors. The learnable distance measure captures motif similarity and domain-specific semantic information such as rotation invariance. The learned distance provides a measurement of semantic similarity between a pair of accelerometer time-series segments, which is used to measure the distance between an anchor and various other sampled candidate segments. The self-supervised model is trained on 1 billion segments from 87,376 participants from a large wearables dataset. The model achieves strong performance across multiple downstream tasks, encompassing both classification and regression. To our knowledge, we are the first to show the generalizability of a self-supervised learning model with motion data from wearables across distinct evaluation tasks.
Latent Temporal Flows for Multivariate Analysis of Wearables Data
Oct 14, 2022



Abstract:Increased use of sensor signals from wearable devices as rich sources of physiological data has sparked growing interest in developing health monitoring systems to identify changes in an individual's health profile. Indeed, machine learning models for sensor signals have enabled a diverse range of healthcare related applications including early detection of abnormalities, fertility tracking, and adverse drug effect prediction. However, these models can fail to account for the dependent high-dimensional nature of the underlying sensor signals. In this paper, we introduce Latent Temporal Flows, a method for multivariate time-series modeling tailored to this setting. We assume that a set of sequences is generated from a multivariate probabilistic model of an unobserved time-varying low-dimensional latent vector. Latent Temporal Flows simultaneously recovers a transformation of the observed sequences into lower-dimensional latent representations via deep autoencoder mappings, and estimates a temporally-conditioned probabilistic model via normalizing flows. Using data from the Apple Heart and Movement Study (AH&MS), we illustrate promising forecasting performance on these challenging signals. Additionally, by analyzing two and three dimensional representations learned by our model, we show that we can identify participants' $\text{VO}_2\text{max}$, a main indicator and summary of cardio-respiratory fitness, using only lower-level signals. Finally, we show that the proposed method consistently outperforms the state-of-the-art in multi-step forecasting benchmarks (achieving at least a $10\%$ performance improvement) on several real-world datasets, while enjoying increased computational efficiency.
PhysioMTL: Personalizing Physiological Patterns using Optimal Transport Multi-Task Regression
Mar 19, 2022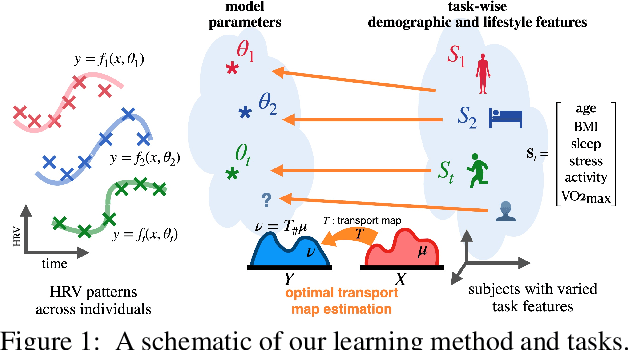
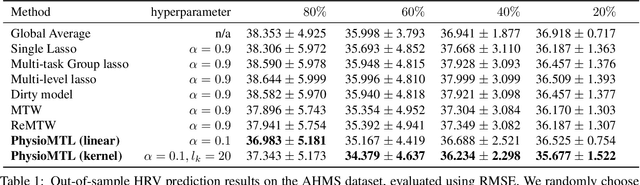
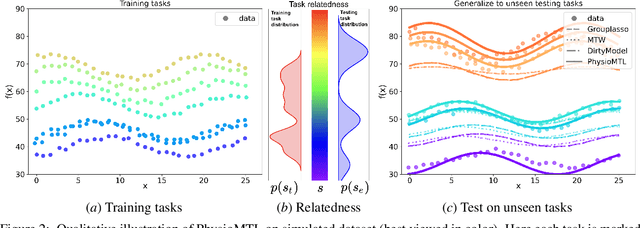
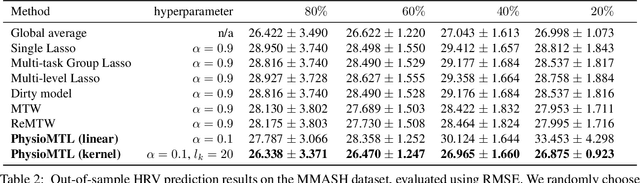
Abstract:Heart rate variability (HRV) is a practical and noninvasive measure of autonomic nervous system activity, which plays an essential role in cardiovascular health. However, using HRV to assess physiology status is challenging. Even in clinical settings, HRV is sensitive to acute stressors such as physical activity, mental stress, hydration, alcohol, and sleep. Wearable devices provide convenient HRV measurements, but the irregularity of measurements and uncaptured stressors can bias conventional analytical methods. To better interpret HRV measurements for downstream healthcare applications, we learn a personalized diurnal rhythm as an accurate physiological indicator for each individual. We develop Physiological Multitask-Learning (PhysioMTL) by harnessing Optimal Transport theory within a Multitask-learning (MTL) framework. The proposed method learns an individual-specific predictive model from heterogeneous observations, and enables estimation of an optimal transport map that yields a push forward operation onto the demographic features for each task. Our model outperforms competing MTL methodologies on unobserved predictive tasks for synthetic and two real-world datasets. Specifically, our method provides remarkable prediction results on unseen held-out subjects given only $20\%$ of the subjects in real-world observational studies. Furthermore, our model enables a counterfactual engine that generates the effect of acute stressors and chronic conditions on HRV rhythms.
Generalizing Variational Autoencoders with Hierarchical Empirical Bayes
Jul 20, 2020



Abstract:Variational Autoencoders (VAEs) have experienced recent success as data-generating models by using simple architectures that do not require significant fine-tuning of hyperparameters. However, VAEs are known to suffer from over-regularization which can lead to failure to escape local maxima. This phenomenon, known as posterior collapse, prevents learning a meaningful latent encoding of the data. Recent methods have mitigated this issue by deterministically moment-matching an aggregated posterior distribution to an aggregate prior. However, abandoning a probabilistic framework (and thus relying on point estimates) can both lead to a discontinuous latent space and generate unrealistic samples. Here we present Hierarchical Empirical Bayes Autoencoder (HEBAE), a computationally stable framework for probabilistic generative models. Our key contributions are two-fold. First, we make gains by placing a hierarchical prior over the encoding distribution, enabling us to adaptively balance the trade-off between minimizing the reconstruction loss function and avoiding over-regularization. Second, we show that assuming a general dependency structure between variables in the latent space produces better convergence onto the mean-field assumption for improved posterior inference. Overall, HEBAE is more robust to a wide-range of hyperparameter initializations than an analogous VAE. Using data from MNIST and CelebA, we illustrate the ability of HEBAE to generate higher quality samples based on FID score than existing autoencoder-based approaches.
Sparse Multi-Output Gaussian Processes for Medical Time Series Prediction
Jun 21, 2018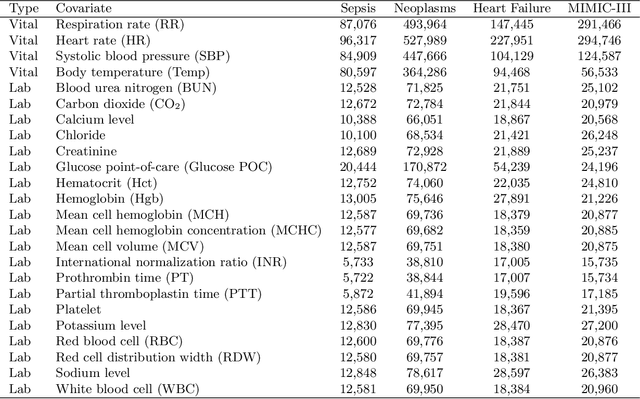
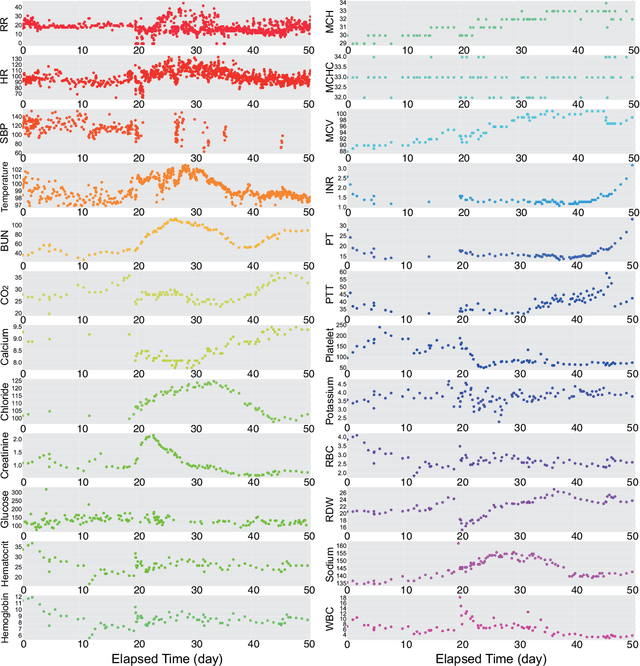
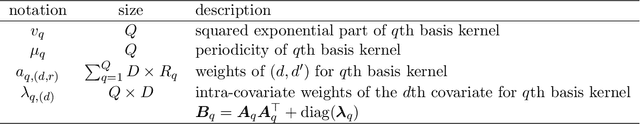
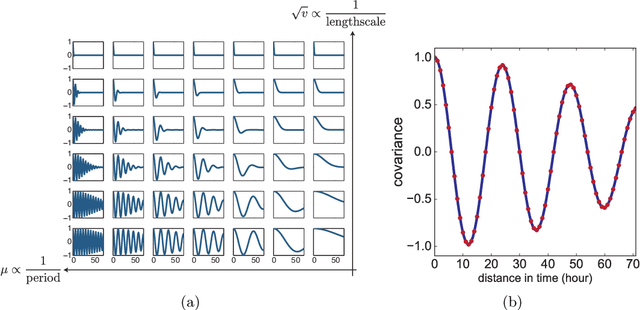
Abstract:In the scenario of real-time monitoring of hospital patients, high-quality inference of patients' health status using all information available from clinical covariates and lab tests is essential to enable successful medical interventions and improve patient outcomes. Developing a computational framework that can learn from observational large-scale electronic health records (EHRs) and make accurate real-time predictions is a critical step. In this work, we develop and explore a Bayesian nonparametric model based on Gaussian process (GP) regression for hospital patient monitoring. We propose MedGP, a statistical framework that incorporates 24 clinical and lab covariates and supports a rich reference data set from which relationships between observed covariates may be inferred and exploited for high-quality inference of patient state over time. To do this, we develop a highly structured sparse GP kernel to enable tractable computation over tens of thousands of time points while estimating correlations among clinical covariates, patients, and periodicity in patient observations. MedGP has a number of benefits over current methods, including (i) not requiring an alignment of the time series data, (ii) quantifying confidence regions in the predictions, (iii) exploiting a vast and rich database of patients, and (iv) inferring interpretable relationships among clinical covariates. We evaluate and compare results from MedGP on the task of online prediction for three patient subgroups from two medical data sets across 8,043 patients. We found MedGP improves online prediction over baseline methods for nearly all covariates across different disease subgroups and studies. The publicly available code is at https://github.com/bee-hive/MedGP.
Adaptive Randomized Dimension Reduction on Massive Data
Apr 13, 2015



Abstract:The scalability of statistical estimators is of increasing importance in modern applications. One approach to implementing scalable algorithms is to compress data into a low dimensional latent space using dimension reduction methods. In this paper we develop an approach for dimension reduction that exploits the assumption of low rank structure in high dimensional data to gain both computational and statistical advantages. We adapt recent randomized low-rank approximation algorithms to provide an efficient solution to principal component analysis (PCA), and we use this efficient solver to improve parameter estimation in large-scale linear mixed models (LMM) for association mapping in statistical and quantitative genomics. A key observation in this paper is that randomization serves a dual role, improving both computational and statistical performance by implicitly regularizing the covariance matrix estimate of the random effect in a LMM. These statistical and computational advantages are highlighted in our experiments on simulated data and large-scale genomic studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge