Fereshteh Shakeri
Histopath-C: Towards Realistic Domain Shifts for Histopathology Vision-Language Adaptation
Jan 18, 2026Abstract:Medical Vision-language models (VLMs) have shown remarkable performances in various medical imaging domains such as histo\-pathology by leveraging pre-trained, contrastive models that exploit visual and textual information. However, histopathology images may exhibit severe domain shifts, such as staining, contamination, blurring, and noise, which may severely degrade the VLM's downstream performance. In this work, we introduce Histopath-C, a new benchmark with realistic synthetic corruptions designed to mimic real-world distribution shifts observed in digital histopathology. Our framework dynamically applies corruptions to any available dataset and evaluates Test-Time Adaptation (TTA) mechanisms on the fly. We then propose LATTE, a transductive, low-rank adaptation strategy that exploits multiple text templates, mitigating the sensitivity of histopathology VLMs to diverse text inputs. Our approach outperforms state-of-the-art TTA methods originally designed for natural images across a breadth of histopathology datasets, demonstrating the effectiveness of our proposed design for robust adaptation in histopathology images. Code and data are available at https://github.com/Mehrdad-Noori/Histopath-C.
UNEM: UNrolled Generalized EM for Transductive Few-Shot Learning
Dec 21, 2024



Abstract:Transductive few-shot learning has recently triggered wide attention in computer vision. Yet, current methods introduce key hyper-parameters, which control the prediction statistics of the test batches, such as the level of class balance, affecting performances significantly. Such hyper-parameters are empirically grid-searched over validation data, and their configurations may vary substantially with the target dataset and pre-training model, making such empirical searches both sub-optimal and computationally intractable. In this work, we advocate and introduce the unrolling paradigm, also referred to as "learning to optimize", in the context of few-shot learning, thereby learning efficiently and effectively a set of optimized hyper-parameters. Specifically, we unroll a generalization of the ubiquitous Expectation-Maximization (EM) optimizer into a neural network architecture, mapping each of its iterates to a layer and learning a set of key hyper-parameters over validation data. Our unrolling approach covers various statistical feature distributions and pre-training paradigms, including recent foundational vision-language models and standard vision-only classifiers. We report comprehensive experiments, which cover a breadth of fine-grained downstream image classification tasks, showing significant gains brought by the proposed unrolled EM algorithm over iterative variants. The achieved improvements reach up to 10% and 7.5% on vision-only and vision-language benchmarks, respectively.
Few-shot Adaptation of Medical Vision-Language Models
Sep 05, 2024Abstract:Integrating image and text data through multi-modal learning has emerged as a new approach in medical imaging research, following its successful deployment in computer vision. While considerable efforts have been dedicated to establishing medical foundation models and their zero-shot transfer to downstream tasks, the popular few-shot setting remains relatively unexplored. Following on from the currently strong emergence of this setting in computer vision, we introduce the first structured benchmark for adapting medical vision-language models (VLMs) in a strict few-shot regime and investigate various adaptation strategies commonly used in the context of natural images. Furthermore, we evaluate a simple generalization of the linear-probe adaptation baseline, which seeks an optimal blending of the visual prototypes and text embeddings via learnable class-wise multipliers. Surprisingly, such a text-informed linear probe yields competitive performances in comparison to convoluted prompt-learning and adapter-based strategies, while running considerably faster and accommodating the black-box setting. Our extensive experiments span three different medical modalities and specialized foundation models, nine downstream tasks, and several state-of-the-art few-shot adaptation methods. We made our benchmark and code publicly available to trigger further developments in this emergent subject: \url{https://github.com/FereshteShakeri/few-shot-MedVLMs}.
Boosting Vision-Language Models for Histopathology Classification: Predict all at once
Sep 03, 2024Abstract:The development of vision-language models (VLMs) for histo-pathology has shown promising new usages and zero-shot performances. However, current approaches, which decompose large slides into smaller patches, focus solely on inductive classification, i.e., prediction for each patch is made independently of the other patches in the target test data. We extend the capability of these large models by introducing a transductive approach. By using text-based predictions and affinity relationships among patches, our approach leverages the strong zero-shot capabilities of these new VLMs without any additional labels. Our experiments cover four histopathology datasets and five different VLMs. Operating solely in the embedding space (i.e., in a black-box setting), our approach is highly efficient, processing $10^5$ patches in just a few seconds, and shows significant accuracy improvements over inductive zero-shot classification. Code available at https://github.com/FereshteShakeri/Histo-TransCLIP.
LP++: A Surprisingly Strong Linear Probe for Few-Shot CLIP
Apr 02, 2024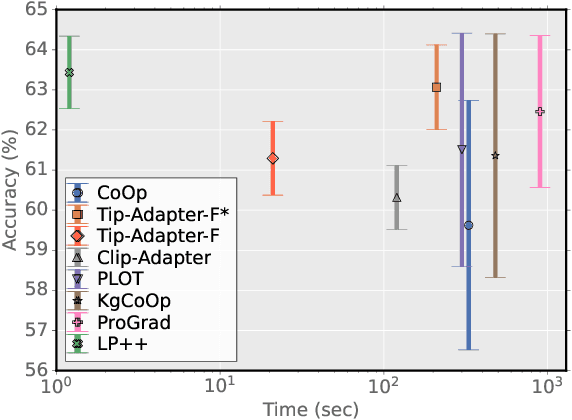
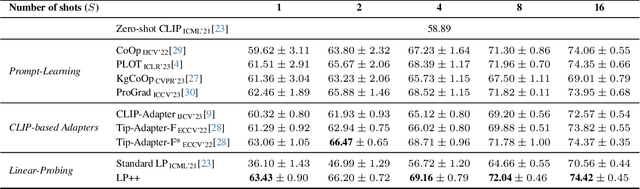
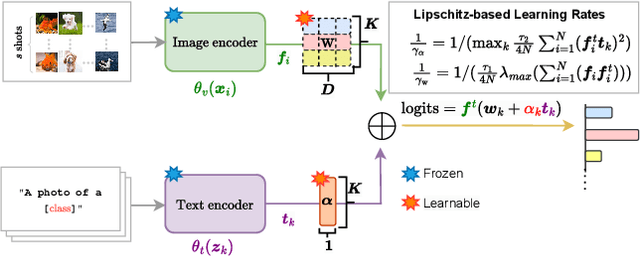
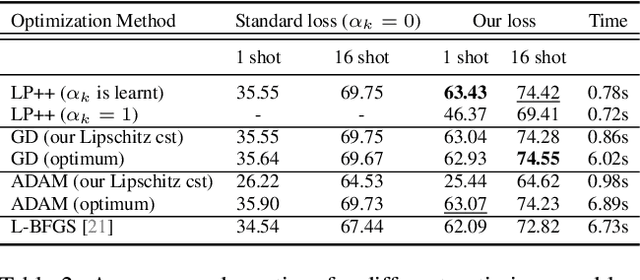
Abstract:In a recent, strongly emergent literature on few-shot CLIP adaptation, Linear Probe (LP) has been often reported as a weak baseline. This has motivated intensive research building convoluted prompt learning or feature adaptation strategies. In this work, we propose and examine from convex-optimization perspectives a generalization of the standard LP baseline, in which the linear classifier weights are learnable functions of the text embedding, with class-wise multipliers blending image and text knowledge. As our objective function depends on two types of variables, i.e., the class visual prototypes and the learnable blending parameters, we propose a computationally efficient block coordinate Majorize-Minimize (MM) descent algorithm. In our full-batch MM optimizer, which we coin LP++, step sizes are implicit, unlike standard gradient descent practices where learning rates are intensively searched over validation sets. By examining the mathematical properties of our loss (e.g., Lipschitz gradient continuity), we build majorizing functions yielding data-driven learning rates and derive approximations of the loss's minima, which provide data-informed initialization of the variables. Our image-language objective function, along with these non-trivial optimization insights and ingredients, yields, surprisingly, highly competitive few-shot CLIP performances. Furthermore, LP++ operates in black-box, relaxes intensive validation searches for the optimization hyper-parameters, and runs orders-of-magnitudes faster than state-of-the-art few-shot CLIP adaptation methods. Our code is available at: \url{https://github.com/FereshteShakeri/FewShot-CLIP-Strong-Baseline.git}.
FHIST: A Benchmark for Few-shot Classification of Histological Images
May 31, 2022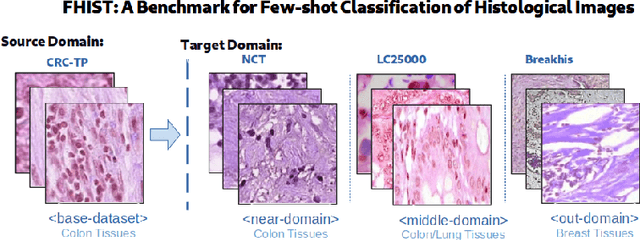
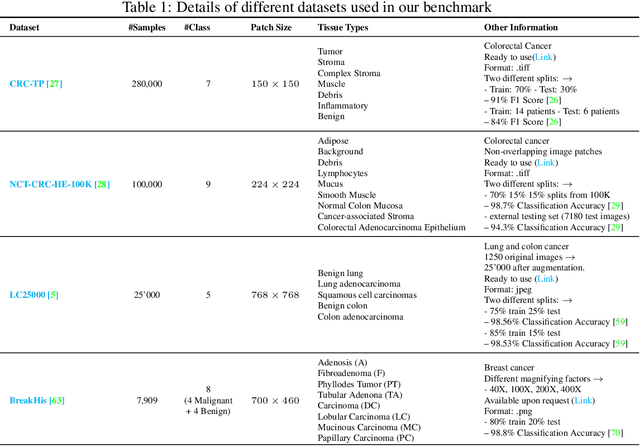
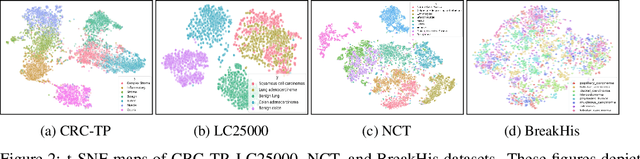
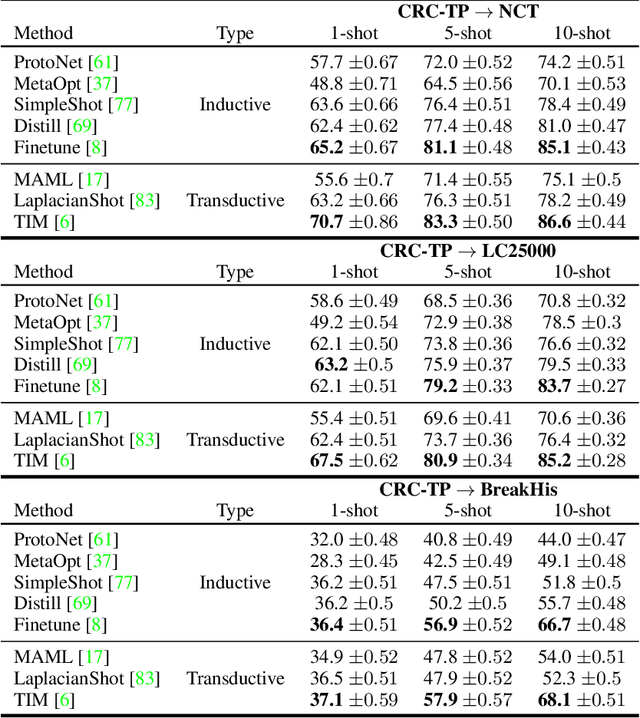
Abstract:Few-shot learning has recently attracted wide interest in image classification, but almost all the current public benchmarks are focused on natural images. The few-shot paradigm is highly relevant in medical-imaging applications due to the scarcity of labeled data, as annotations are expensive and require specialized expertise. However, in medical imaging, few-shot learning research is sparse, limited to private data sets and is at its early stage. In particular, the few-shot setting is of high interest in histology due to the diversity and fine granularity of cancer related tissue classification tasks, and the variety of data-preparation techniques. This paper introduces a highly diversified public benchmark, gathered from various public datasets, for few-shot histology data classification. We build few-shot tasks and base-training data with various tissue types, different levels of domain shifts stemming from various cancer sites, and different class-granularity levels, thereby reflecting realistic scenarios. We evaluate the performances of state-of-the-art few-shot learning methods on our benchmark, and observe that simple fine-tuning and regularization methods achieve better results than the popular meta-learning and episodic-training paradigm. Furthermore, we introduce three scenarios based on the domain shifts between the source and target histology data: near-domain, middle-domain and out-domain. Our experiments display the potential of few-shot learning in histology classification, with state-of-art few shot learning methods approaching the supervised-learning baselines in the near-domain setting. In our out-domain setting, for 5-way 5-shot, the best performing method reaches 60% accuracy. We believe that our work could help in building realistic evaluations and fair comparisons of few-shot learning methods and will further encourage research in the few-shot paradigm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge