Aymen Sadraoui
OPIS, CVN
UNEM: UNrolled Generalized EM for Transductive Few-Shot Learning
Dec 21, 2024



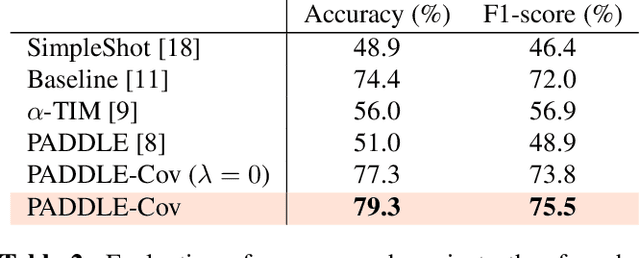
Abstract:Transductive few-shot learning has recently triggered wide attention in computer vision. Yet, current methods introduce key hyper-parameters, which control the prediction statistics of the test batches, such as the level of class balance, affecting performances significantly. Such hyper-parameters are empirically grid-searched over validation data, and their configurations may vary substantially with the target dataset and pre-training model, making such empirical searches both sub-optimal and computationally intractable. In this work, we advocate and introduce the unrolling paradigm, also referred to as "learning to optimize", in the context of few-shot learning, thereby learning efficiently and effectively a set of optimized hyper-parameters. Specifically, we unroll a generalization of the ubiquitous Expectation-Maximization (EM) optimizer into a neural network architecture, mapping each of its iterates to a layer and learning a set of key hyper-parameters over validation data. Our unrolling approach covers various statistical feature distributions and pre-training paradigms, including recent foundational vision-language models and standard vision-only classifiers. We report comprehensive experiments, which cover a breadth of fine-grained downstream image classification tasks, showing significant gains brought by the proposed unrolled EM algorithm over iterative variants. The achieved improvements reach up to 10% and 7.5% on vision-only and vision-language benchmarks, respectively.
A transductive few-shot learning approach for classification of digital histopathological slides from liver cancer
Nov 29, 2023
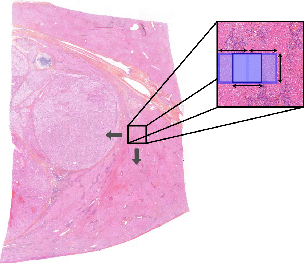
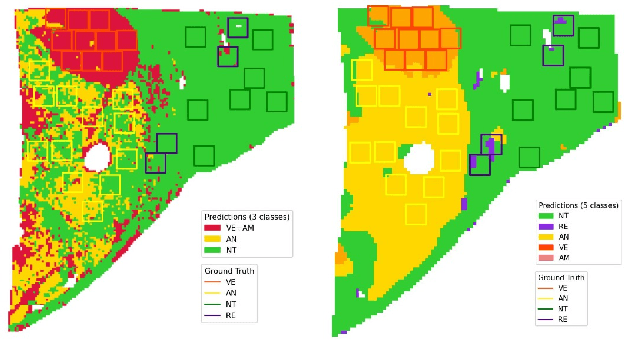
Abstract:This paper presents a new approach for classifying 2D histopathology patches using few-shot learning. The method is designed to tackle a significant challenge in histopathology, which is the limited availability of labeled data. By applying a sliding window technique to histopathology slides, we illustrate the practical benefits of transductive learning (i.e., making joint predictions on patches) to achieve consistent and accurate classification. Our approach involves an optimization-based strategy that actively penalizes the prediction of a large number of distinct classes within each window. We conducted experiments on histopathological data to classify tissue classes in digital slides of liver cancer, specifically hepatocellular carcinoma. The initial results show the effectiveness of our method and its potential to enhance the process of automated cancer diagnosis and treatment, all while reducing the time and effort required for expert annotation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge