Femke Ongenae
From Haystack to Needle: Label Space Reduction for Zero-shot Classification
Feb 12, 2025Abstract:We present Label Space Reduction (LSR), a novel method for improving zero-shot classification performance of Large Language Models (LLMs). LSR iteratively refines the classification label space by systematically ranking and reducing candidate classes, enabling the model to concentrate on the most relevant options. By leveraging unlabeled data with the statistical learning capabilities of data-driven models, LSR dynamically optimizes the label space representation at test time. Our experiments across seven benchmarks demonstrate that LSR improves macro-F1 scores by an average of 7.0% (up to 14.2%) with Llama-3.1-70B and 3.3% (up to 11.1%) with Claude-3.5-Sonnet compared to standard zero-shot classification baselines. To reduce the computational overhead of LSR, which requires an additional LLM call at each iteration, we propose distilling the model into a probabilistic classifier, allowing for efficient inference.
Perfectly predicting ICU length of stay: too good to be true
Nov 10, 2022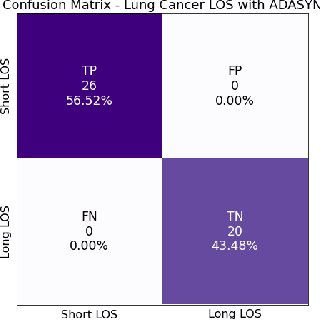
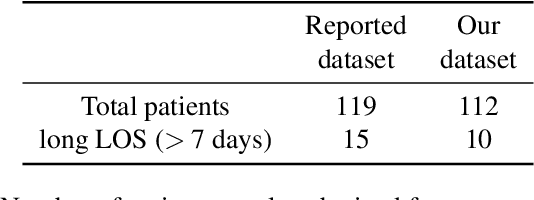

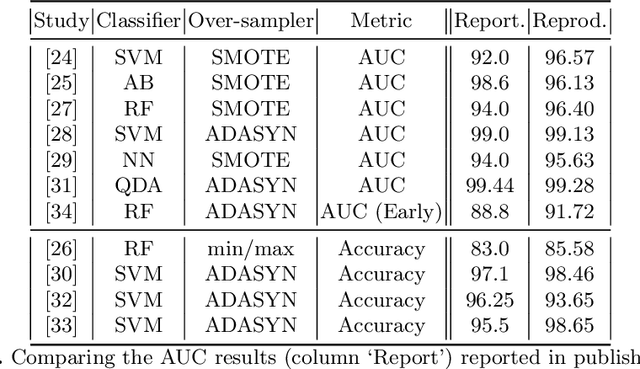
Abstract:A paper of Alsinglawi et al was recently accepted and published in Scientific Reports. In this paper, the authors aim to predict length of stay (LOS), discretized into either long (> 7 days) or short stays (< 7 days), of lung cancer patients in an ICU department using various machine learning techniques. The authors claim to achieve perfect results with an Area Under the Receiver Operating Characteristic curve (AUROC) of 100% with a Random Forest (RF) classifier with ADASYN class balancing over sampling technique, which if accurate could have significant implications for hospital management. However, we have identified several methodological flaws within the manuscript which cause the results to be overly optimistic and would have serious consequences if used in a clinical practice. Moreover, the reporting of the methodology is unclear and many important details are missing from the manuscript, which makes reproduction extremely difficult. We highlight the effect these oversights have had on the result and provide a more believable result of 88.91% AUROC when these oversights are corrected.
Powershap: A Power-full Shapley Feature Selection Method
Jun 16, 2022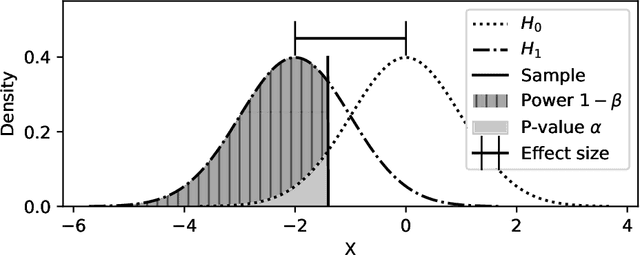
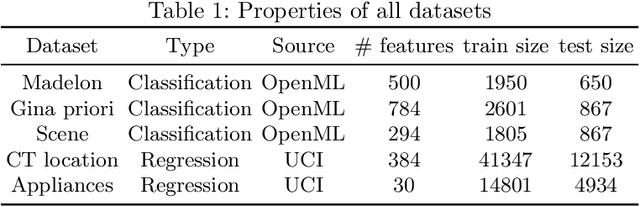
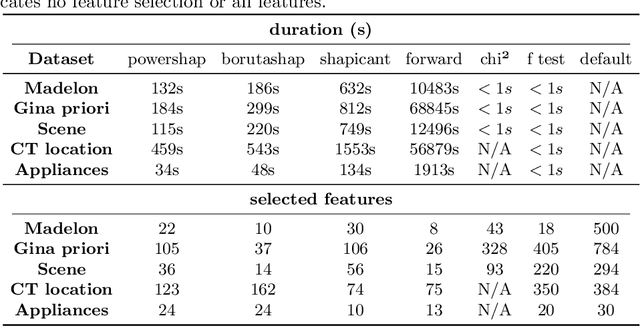

Abstract:Feature selection is a crucial step in developing robust and powerful machine learning models. Feature selection techniques can be divided into two categories: filter and wrapper methods. While wrapper methods commonly result in strong predictive performances, they suffer from a large computational complexity and therefore take a significant amount of time to complete, especially when dealing with high-dimensional feature sets. Alternatively, filter methods are considerably faster, but suffer from several other disadvantages, such as (i) requiring a threshold value, (ii) not taking into account intercorrelation between features, and (iii) ignoring feature interactions with the model. To this end, we present powershap, a novel wrapper feature selection method, which leverages statistical hypothesis testing and power calculations in combination with Shapley values for quick and intuitive feature selection. Powershap is built on the core assumption that an informative feature will have a larger impact on the prediction compared to a known random feature. Benchmarks and simulations show that powershap outperforms other filter methods with predictive performances on par with wrapper methods while being significantly faster, often even reaching half or a third of the execution time. As such, powershap provides a competitive and quick algorithm that can be used by various models in different domains. Furthermore, powershap is implemented as a plug-and-play and open-source sklearn component, enabling easy integration in conventional data science pipelines. User experience is even further enhanced by also providing an automatic mode that automatically tunes the hyper-parameters of the powershap algorithm, allowing to use the algorithm without any configuration needed.
pyRDF2Vec: A Python Implementation and Extension of RDF2Vec
May 04, 2022
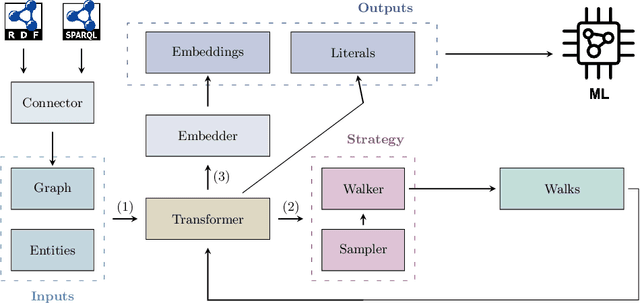
Abstract:This paper introduces pyRDF2Vec, a Python software package that reimplements the well-known RDF2Vec algorithm along with several of its extensions. By making the algorithm available in the most popular data science language, and by bundling all extensions into a single place, the use of RDF2Vec is simplified for data scientists. The package is released under a MIT license and structured in such a way to foster further research into sampling, walking, and embedding strategies, which are vital components of the RDF2Vec algorithm. Several optimisations have been implemented in \texttt{pyRDF2Vec} that allow for more efficient walk extraction than the original algorithm. Furthermore, best practices in terms of code styling, testing, and documentation were applied such that the package is future-proof as well as to facilitate external contributions.
R-GCN: The R Could Stand for Random
Mar 04, 2022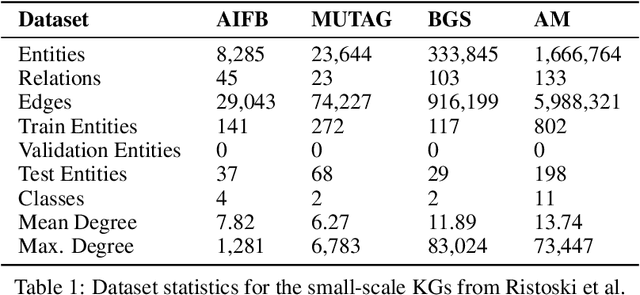
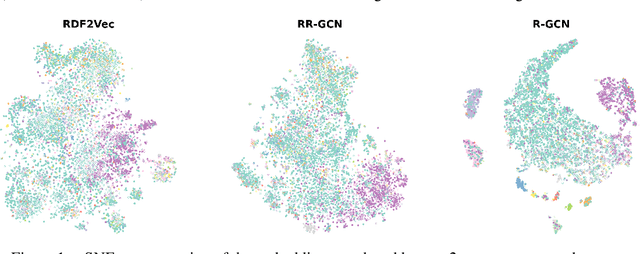
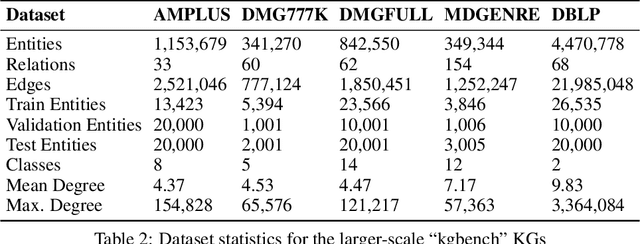
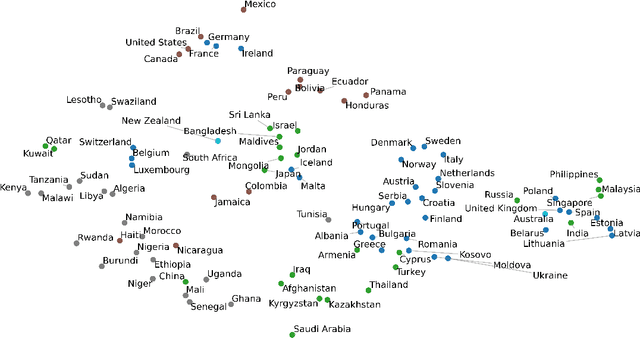
Abstract:The inception of Relational Graph Convolutional Networks (R-GCNs) marked a milestone in the Semantic Web domain as it allows for end-to-end training of machine learning models that operate on Knowledge Graphs (KGs). R-GCNs generate a representation for a node of interest by repeatedly aggregating parametrised, relation-specific transformations of its neighbours. However, in this paper, we argue that the the R-GCN's main contribution lies in this "message passing" paradigm, rather than the learned parameters. To this end, we introduce the "Random Relational Graph Convolutional Network" (RR-GCN), which constructs embeddings for nodes in the KG by aggregating randomly transformed random information from neigbours, i.e., with no learned parameters. We empirically show that RR-GCNs can compete with fully trained R-GCNs in both node classification and link prediction settings. The implications of these results are two-fold: on the one hand, our technique can be used as a quick baseline that novel KG embedding methods should be able to beat. On the other hand, it demonstrates that further research might reveal more parameter-efficient inductive biases for KGs.
Walk Extraction Strategies for Node Embeddings with RDF2Vec in Knowledge Graphs
Sep 09, 2020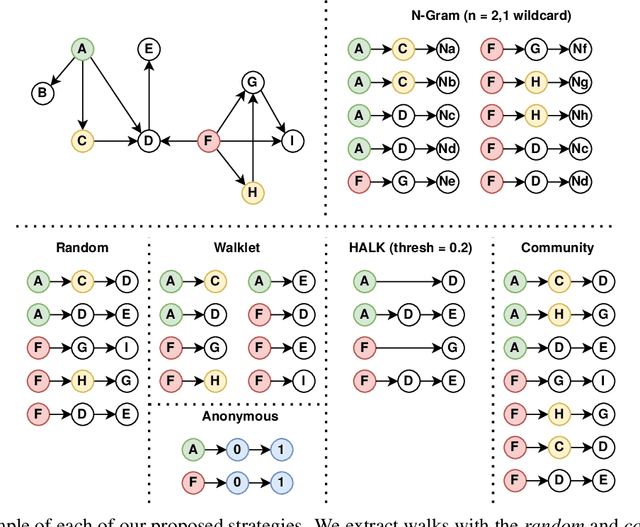
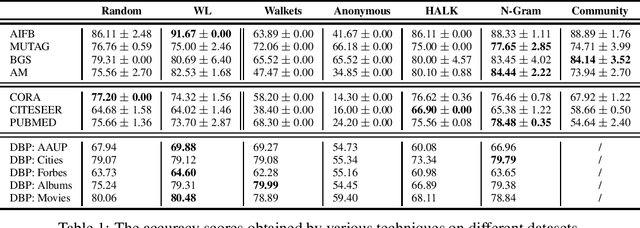

Abstract:As KGs are symbolic constructs, specialized techniques have to be applied in order to make them compatible with data mining techniques. RDF2Vec is an unsupervised technique that can create task-agnostic numerical representations of the nodes in a KG by extending successful language modelling techniques. The original work proposed the Weisfeiler-Lehman (WL) kernel to improve the quality of the representations. However, in this work, we show both formally and empirically that the WL kernel does little to improve walk embeddings in the context of a single KG. As an alternative to the WL kernel, we propose five different strategies to extract information complementary to basic random walks. We compare these walks on several benchmark datasets to show that the \emph{n-gram} strategy performs best on average on node classification tasks and that tuning the walk strategy can result in improved predictive performances.
Overly Optimistic Prediction Results on Imbalanced Data: Flaws and Benefits of Applying Over-sampling
Jan 15, 2020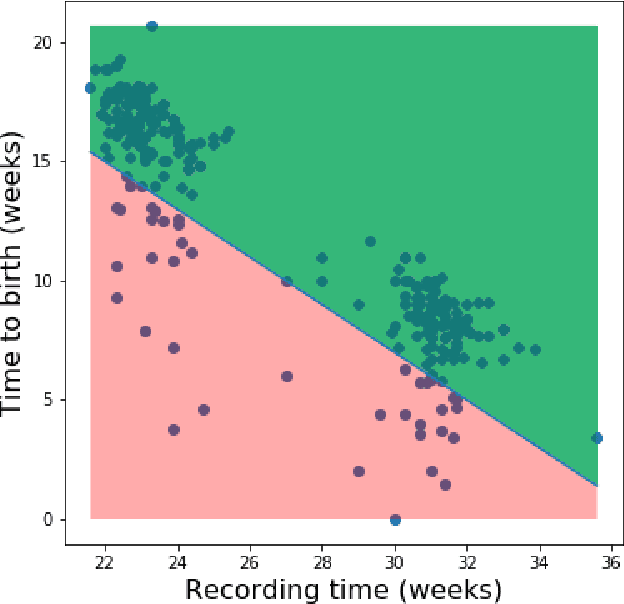
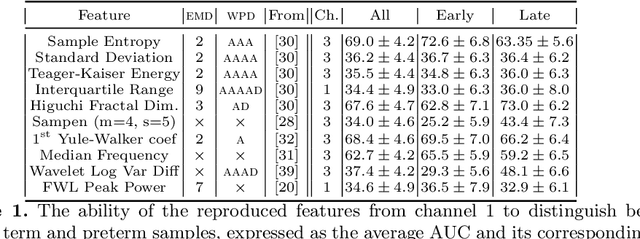
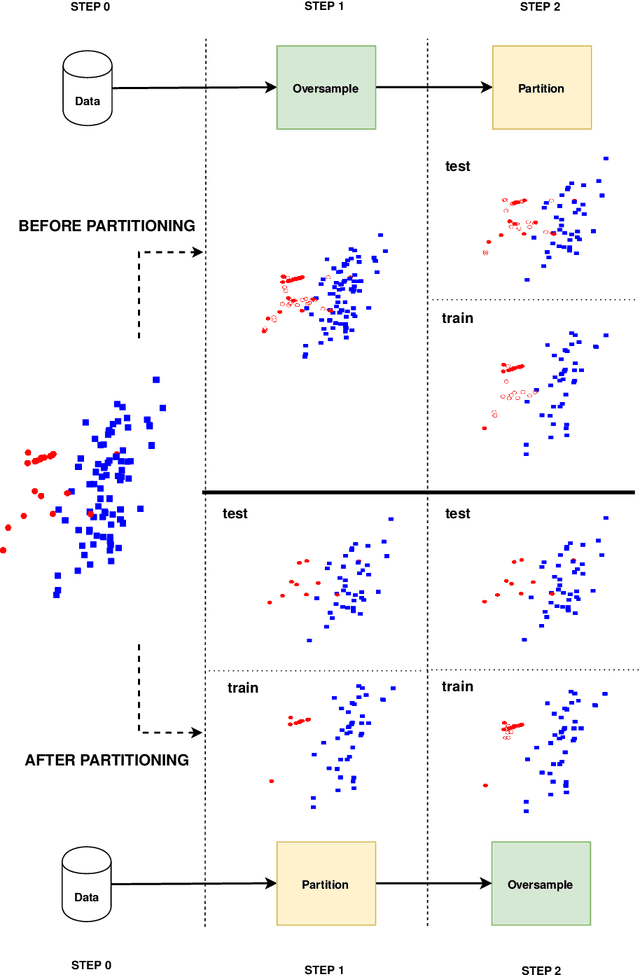
Abstract:Information extracted from electrohysterography recordings could potentially prove to be an interesting additional source of information to estimate the risk on preterm birth. Recently, a large number of studies have reported near-perfect results to distinguish between recordings of patients that will deliver term or preterm using a public resource, called the Term/Preterm Electrohysterogram database. However, we argue that these results are overly optimistic due to a methodological flaw being made. In this work, we focus on one specific type of methodological flaw: applying over-sampling before partitioning the data into mutually exclusive training and testing sets. We show how this causes the results to be biased using two artificial datasets and reproduce results of studies in which this flaw was identified. Moreover, we evaluate the actual impact of over-sampling on predictive performance, when applied prior to data partitioning, using the same methodologies of related studies, to provide a realistic view of these methodologies' generalization capabilities. We make our research reproducible by providing all the code under an open license.
Positive blood culture detection in time series data using a BiLSTM network
Dec 03, 2016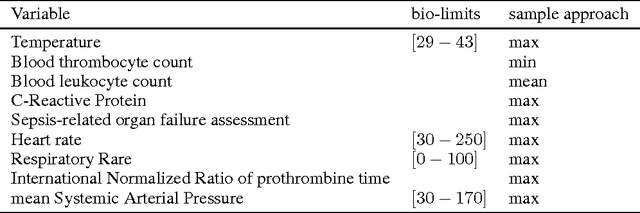


Abstract:The presence of bacteria or fungi in the bloodstream of patients is abnormal and can lead to life-threatening conditions. A computational model based on a bidirectional long short-term memory artificial neural network, is explored to assist doctors in the intensive care unit to predict whether examination of blood cultures of patients will return positive. As input it uses nine monitored clinical parameters, presented as time series data, collected from 2177 ICU admissions at the Ghent University Hospital. Our main goal is to determine if general machine learning methods and more specific, temporal models, can be used to create an early detection system. This preliminary research obtains an area of 71.95% under the precision recall curve, proving the potential of temporal neural networks in this context.
GENESIM: genetic extraction of a single, interpretable model
Nov 17, 2016

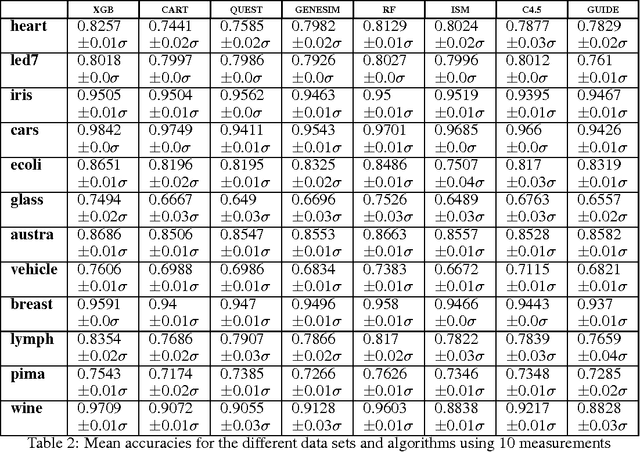
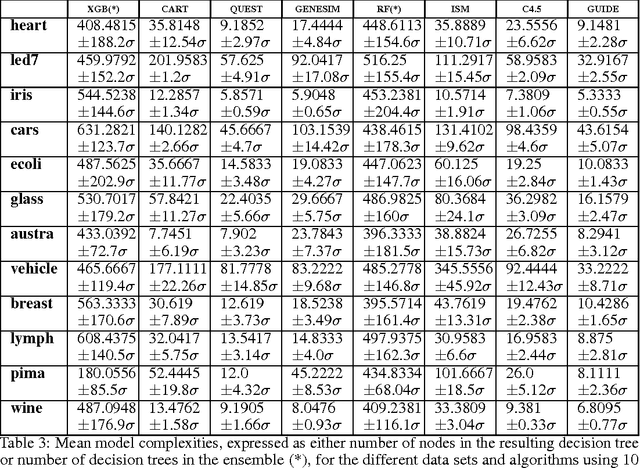
Abstract:Models obtained by decision tree induction techniques excel in being interpretable.However, they can be prone to overfitting, which results in a low predictive performance. Ensemble techniques are able to achieve a higher accuracy. However, this comes at a cost of losing interpretability of the resulting model. This makes ensemble techniques impractical in applications where decision support, instead of decision making, is crucial. To bridge this gap, we present the GENESIM algorithm that transforms an ensemble of decision trees to a single decision tree with an enhanced predictive performance by using a genetic algorithm. We compared GENESIM to prevalent decision tree induction and ensemble techniques using twelve publicly available data sets. The results show that GENESIM achieves a better predictive performance on most of these data sets than decision tree induction techniques and a predictive performance in the same order of magnitude as the ensemble techniques. Moreover, the resulting model of GENESIM has a very low complexity, making it very interpretable, in contrast to ensemble techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge