Bram Steenwinckel
From Haystack to Needle: Label Space Reduction for Zero-shot Classification
Feb 12, 2025Abstract:We present Label Space Reduction (LSR), a novel method for improving zero-shot classification performance of Large Language Models (LLMs). LSR iteratively refines the classification label space by systematically ranking and reducing candidate classes, enabling the model to concentrate on the most relevant options. By leveraging unlabeled data with the statistical learning capabilities of data-driven models, LSR dynamically optimizes the label space representation at test time. Our experiments across seven benchmarks demonstrate that LSR improves macro-F1 scores by an average of 7.0% (up to 14.2%) with Llama-3.1-70B and 3.3% (up to 11.1%) with Claude-3.5-Sonnet compared to standard zero-shot classification baselines. To reduce the computational overhead of LSR, which requires an additional LLM call at each iteration, we propose distilling the model into a probabilistic classifier, allowing for efficient inference.
pyRDF2Vec: A Python Implementation and Extension of RDF2Vec
May 04, 2022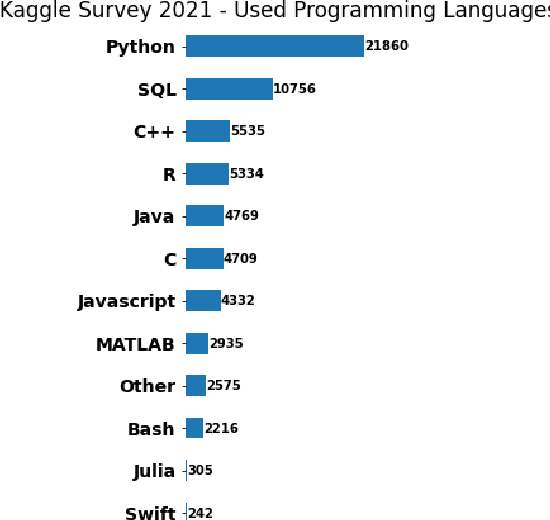
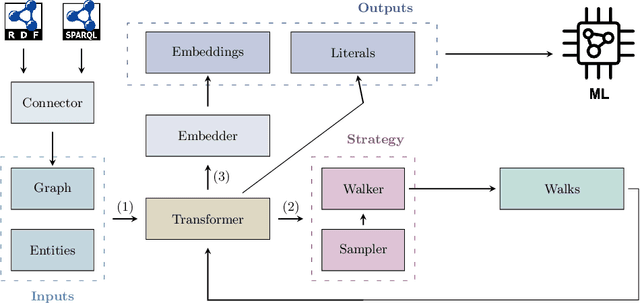
Abstract:This paper introduces pyRDF2Vec, a Python software package that reimplements the well-known RDF2Vec algorithm along with several of its extensions. By making the algorithm available in the most popular data science language, and by bundling all extensions into a single place, the use of RDF2Vec is simplified for data scientists. The package is released under a MIT license and structured in such a way to foster further research into sampling, walking, and embedding strategies, which are vital components of the RDF2Vec algorithm. Several optimisations have been implemented in \texttt{pyRDF2Vec} that allow for more efficient walk extraction than the original algorithm. Furthermore, best practices in terms of code styling, testing, and documentation were applied such that the package is future-proof as well as to facilitate external contributions.
Predictive models of RNA degradation through dual crowdsourcing
Oct 14, 2021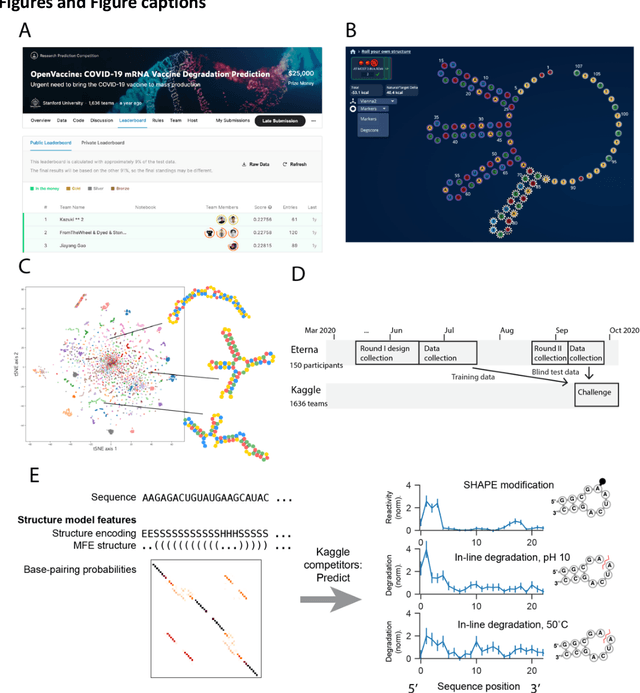
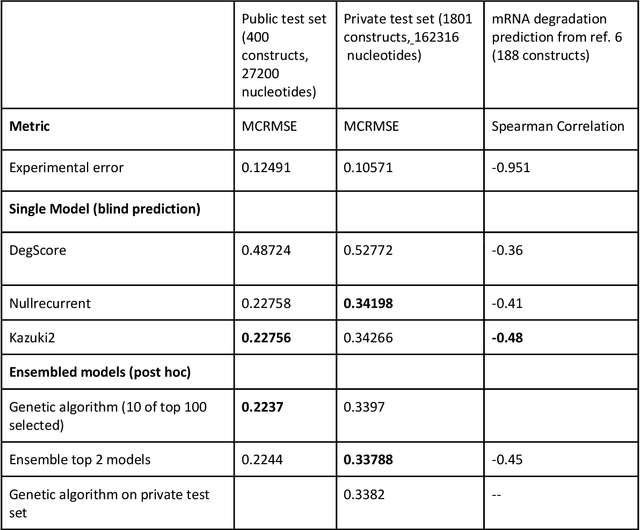
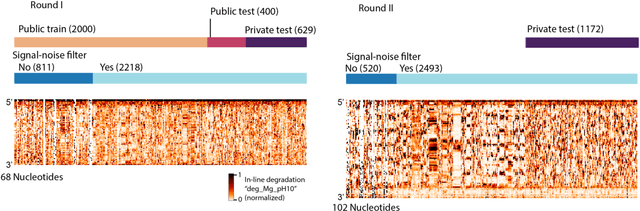
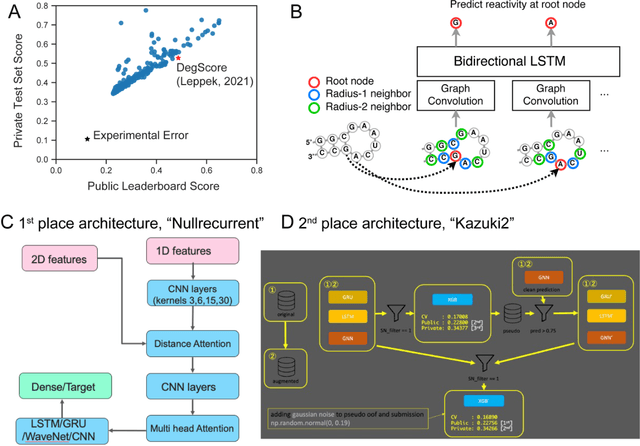
Abstract:Messenger RNA-based medicines hold immense potential, as evidenced by their rapid deployment as COVID-19 vaccines. However, worldwide distribution of mRNA molecules has been limited by their thermostability, which is fundamentally limited by the intrinsic instability of RNA molecules to a chemical degradation reaction called in-line hydrolysis. Predicting the degradation of an RNA molecule is a key task in designing more stable RNA-based therapeutics. Here, we describe a crowdsourced machine learning competition ("Stanford OpenVaccine") on Kaggle, involving single-nucleotide resolution measurements on 6043 102-130-nucleotide diverse RNA constructs that were themselves solicited through crowdsourcing on the RNA design platform Eterna. The entire experiment was completed in less than 6 months. Winning models demonstrated test set errors that were better by 50% than the previous state-of-the-art DegScore model. Furthermore, these models generalized to blindly predicting orthogonal degradation data on much longer mRNA molecules (504-1588 nucleotides) with improved accuracy over DegScore and other models. Top teams integrated natural language processing architectures and data augmentation techniques with predictions from previous dynamic programming models for RNA secondary structure. These results indicate that such models are capable of representing in-line hydrolysis with excellent accuracy, supporting their use for designing stabilized messenger RNAs. The integration of two crowdsourcing platforms, one for data set creation and another for machine learning, may be fruitful for other urgent problems that demand scientific discovery on rapid timescales.
Walk Extraction Strategies for Node Embeddings with RDF2Vec in Knowledge Graphs
Sep 09, 2020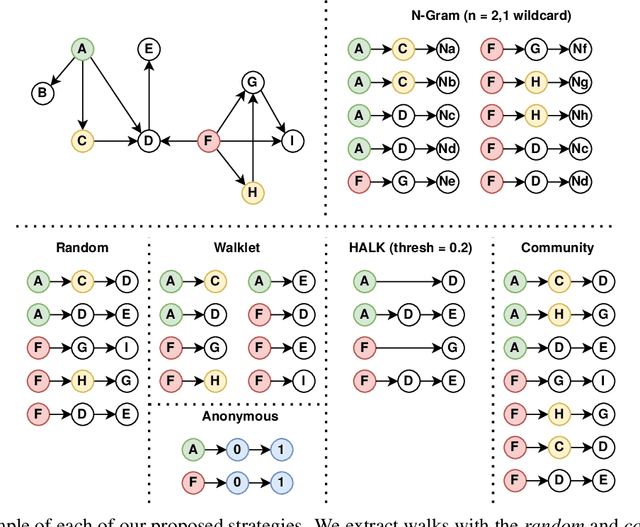
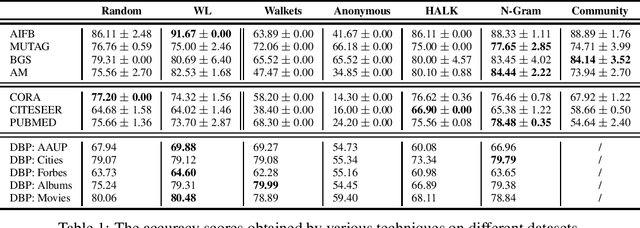

Abstract:As KGs are symbolic constructs, specialized techniques have to be applied in order to make them compatible with data mining techniques. RDF2Vec is an unsupervised technique that can create task-agnostic numerical representations of the nodes in a KG by extending successful language modelling techniques. The original work proposed the Weisfeiler-Lehman (WL) kernel to improve the quality of the representations. However, in this work, we show both formally and empirically that the WL kernel does little to improve walk embeddings in the context of a single KG. As an alternative to the WL kernel, we propose five different strategies to extract information complementary to basic random walks. We compare these walks on several benchmark datasets to show that the \emph{n-gram} strategy performs best on average on node classification tasks and that tuning the walk strategy can result in improved predictive performances.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge