Maggie Demkin
Position: AI Competitions Provide the Gold Standard for Empirical Rigor in GenAI Evaluation
May 01, 2025Abstract:In this position paper, we observe that empirical evaluation in Generative AI is at a crisis point since traditional ML evaluation and benchmarking strategies are insufficient to meet the needs of evaluating modern GenAI models and systems. There are many reasons for this, including the fact that these models typically have nearly unbounded input and output spaces, typically do not have a well defined ground truth target, and typically exhibit strong feedback loops and prediction dependence based on context of previous model outputs. On top of these critical issues, we argue that the problems of {\em leakage} and {\em contamination} are in fact the most important and difficult issues to address for GenAI evaluations. Interestingly, the field of AI Competitions has developed effective measures and practices to combat leakage for the purpose of counteracting cheating by bad actors within a competition setting. This makes AI Competitions an especially valuable (but underutilized) resource. Now is time for the field to view AI Competitions as the gold standard for empirical rigor in GenAI evaluation, and to harness and harvest their results with according value.
Predictive models of RNA degradation through dual crowdsourcing
Oct 14, 2021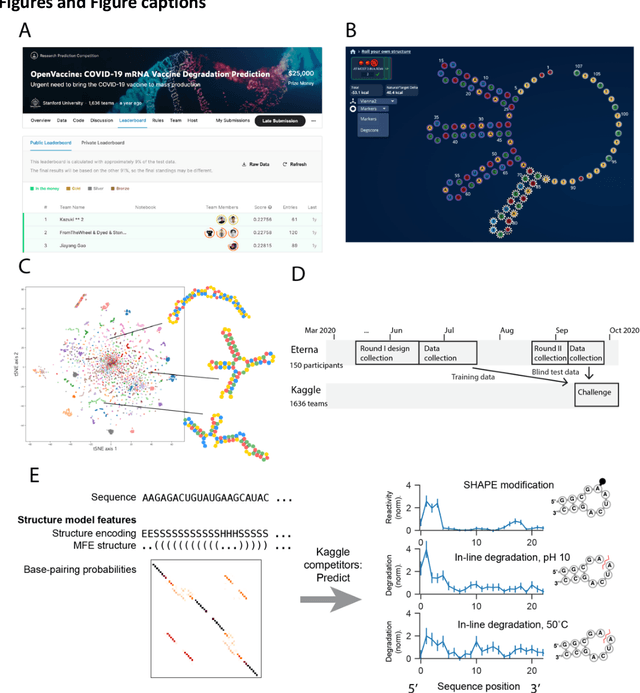
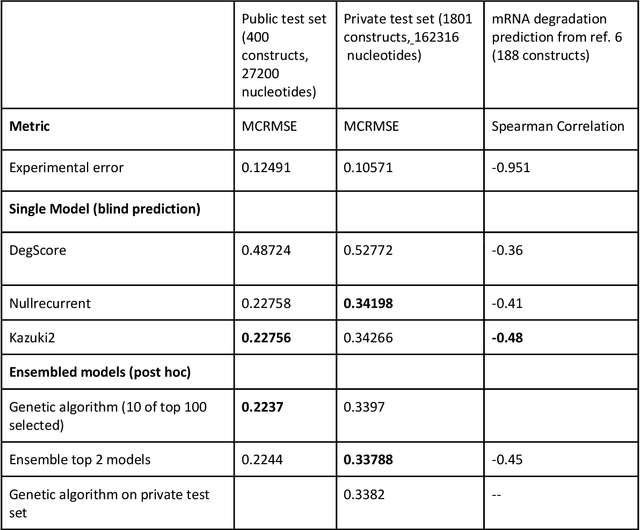
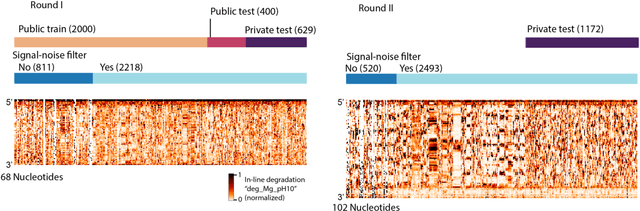
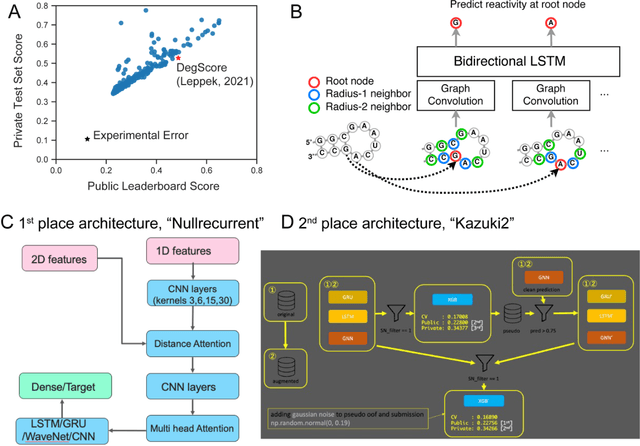
Abstract:Messenger RNA-based medicines hold immense potential, as evidenced by their rapid deployment as COVID-19 vaccines. However, worldwide distribution of mRNA molecules has been limited by their thermostability, which is fundamentally limited by the intrinsic instability of RNA molecules to a chemical degradation reaction called in-line hydrolysis. Predicting the degradation of an RNA molecule is a key task in designing more stable RNA-based therapeutics. Here, we describe a crowdsourced machine learning competition ("Stanford OpenVaccine") on Kaggle, involving single-nucleotide resolution measurements on 6043 102-130-nucleotide diverse RNA constructs that were themselves solicited through crowdsourcing on the RNA design platform Eterna. The entire experiment was completed in less than 6 months. Winning models demonstrated test set errors that were better by 50% than the previous state-of-the-art DegScore model. Furthermore, these models generalized to blindly predicting orthogonal degradation data on much longer mRNA molecules (504-1588 nucleotides) with improved accuracy over DegScore and other models. Top teams integrated natural language processing architectures and data augmentation techniques with predictions from previous dynamic programming models for RNA secondary structure. These results indicate that such models are capable of representing in-line hydrolysis with excellent accuracy, supporting their use for designing stabilized messenger RNAs. The integration of two crowdsourcing platforms, one for data set creation and another for machine learning, may be fruitful for other urgent problems that demand scientific discovery on rapid timescales.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge