Federico Bolelli
Update Your Transformer to the Latest Release: Re-Basin of Task Vectors
May 28, 2025Abstract:Foundation models serve as the backbone for numerous specialized models developed through fine-tuning. However, when the underlying pretrained model is updated or retrained (e.g., on larger and more curated datasets), the fine-tuned model becomes obsolete, losing its utility and requiring retraining. This raises the question: is it possible to transfer fine-tuning to a new release of the model? In this work, we investigate how to transfer fine-tuning to a new checkpoint without having to re-train, in a data-free manner. To do so, we draw principles from model re-basin and provide a recipe based on weight permutations to re-base the modifications made to the original base model, often called task vector. In particular, our approach tailors model re-basin for Transformer models, taking into account the challenges of residual connections and multi-head attention layers. Specifically, we propose a two-level method rooted in spectral theory, initially permuting the attention heads and subsequently adjusting parameters within select pairs of heads. Through extensive experiments on visual and textual tasks, we achieve the seamless transfer of fine-tuned knowledge to new pre-trained backbones without relying on a single training step or datapoint. Code is available at https://github.com/aimagelab/TransFusion.
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
Taming Mambas for Voxel Level 3D Medical Image Segmentation
Oct 20, 2024Abstract:Recently, the field of 3D medical segmentation has been dominated by deep learning models employing Convolutional Neural Networks (CNNs) and Transformer-based architectures, each with their distinctive strengths and limitations. CNNs are constrained by a local receptive field, whereas transformers are hindered by their substantial memory requirements as well as they data hungriness, making them not ideal for processing 3D medical volumes at a fine-grained level. For these reasons, fully convolutional neural networks, as nnUNet, still dominate the scene when segmenting medical structures in 3D large medical volumes. Despite numerous advancements towards developing transformer variants with subquadratic time and memory complexity, these models still fall short in content-based reasoning. A recent breakthrough is Mamba, a Recurrent Neural Network (RNN) based on State Space Models (SSMs) outperforming Transformers in many long-context tasks (million-length sequences) on famous natural language processing and genomic benchmarks while keeping a linear complexity.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
M-VAD Names: a Dataset for Video Captioning with Naming
Mar 04, 2019
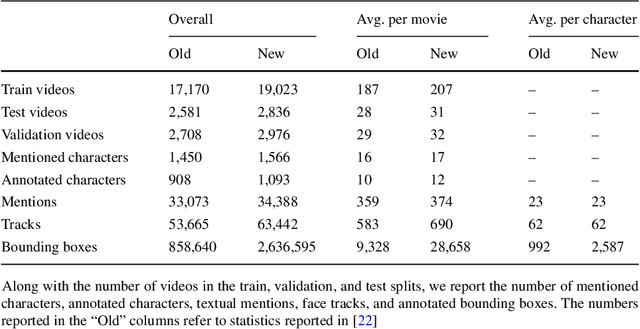
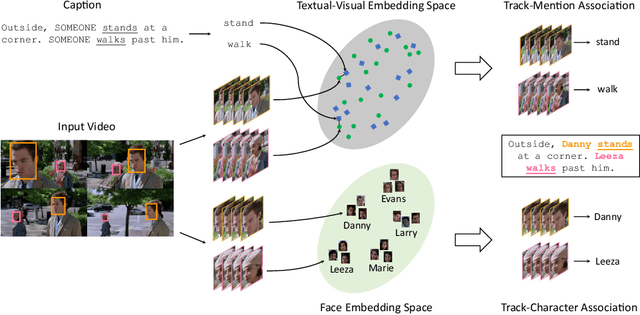
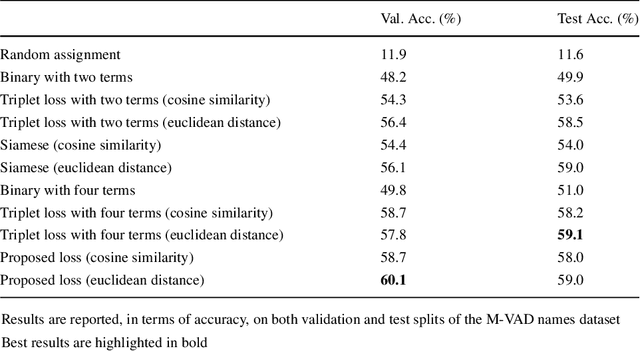
Abstract:Current movie captioning architectures are not capable of mentioning characters with their proper name, replacing them with a generic "someone" tag. The lack of movie description datasets with characters' visual annotations surely plays a relevant role in this shortage. Recently, we proposed to extend the M-VAD dataset by introducing such information. In this paper, we present an improved version of the dataset, namely M-VAD Names, and its semi-automatic annotation procedure. The resulting dataset contains 63k visual tracks and 34k textual mentions, all associated with character identities. To showcase the features of the dataset and quantify the complexity of the naming task, we investigate multimodal architectures to replace the "someone" tags with proper character names in existing video captions. The evaluation is further extended by testing this application on videos outside of the M-VAD Names dataset.
* Source Code: https://github.com/aimagelab/mvad-names-dataset - Video Demo: https://youtu.be/dOvtAXbOOH4
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge