Evgeny M. Mirkes
When fractional quasi p-norms concentrate
May 26, 2025Abstract:Concentration of distances in high dimension is an important factor for the development and design of stable and reliable data analysis algorithms. In this paper, we address the fundamental long-standing question about the concentration of distances in high dimension for fractional quasi $p$-norms, $p\in(0,1)$. The topic has been at the centre of various theoretical and empirical controversies. Here we, for the first time, identify conditions when fractional quasi $p$-norms concentrate and when they don't. We show that contrary to some earlier suggestions, for broad classes of distributions, fractional quasi $p$-norms admit exponential and uniform in $p$ concentration bounds. For these distributions, the results effectively rule out previously proposed approaches to alleviate concentration by "optimal" setting the values of $p$ in $(0,1)$. At the same time, we specify conditions and the corresponding families of distributions for which one can still control concentration rates by appropriate choices of $p$. We also show that in an arbitrarily small vicinity of a distribution from a large class of distributions for which uniform concentration occurs, there are uncountably many other distributions featuring anti-concentration properties. Importantly, this behavior enables devising relevant data encoding or representation schemes favouring or discouraging distance concentration. The results shed new light on this long-standing problem and resolve the tension around the topic in both theory and empirical evidence reported in the literature.
What is Hiding in Medicine's Dark Matter? Learning with Missing Data in Medical Practices
Feb 09, 2024



Abstract:Electronic patient records (EPRs) produce a wealth of data but contain significant missing information. Understanding and handling this missing data is an important part of clinical data analysis and if left unaddressed could result in bias in analysis and distortion in critical conclusions. Missing data may be linked to health care professional practice patterns and imputation of missing data can increase the validity of clinical decisions. This study focuses on statistical approaches for understanding and interpreting the missing data and machine learning based clinical data imputation using a single centre's paediatric emergency data and the data from UK's largest clinical audit for traumatic injury database (TARN). In the study of 56,961 data points related to initial vital signs and observations taken on children presenting to an Emergency Department, we have shown that missing data are likely to be non-random and how these are linked to health care professional practice patterns. We have then examined 79 TARN fields with missing values for 5,791 trauma cases. Singular Value Decomposition (SVD) and k-Nearest Neighbour (kNN) based missing data imputation methods are used and imputation results against the original dataset are compared and statistically tested. We have concluded that the 1NN imputer is the best imputation which indicates a usual pattern of clinical decision making: find the most similar patients and take their attributes as imputation.
* 8 pages
Weakly Supervised Learners for Correction of AI Errors with Provable Performance Guarantees
Feb 06, 2024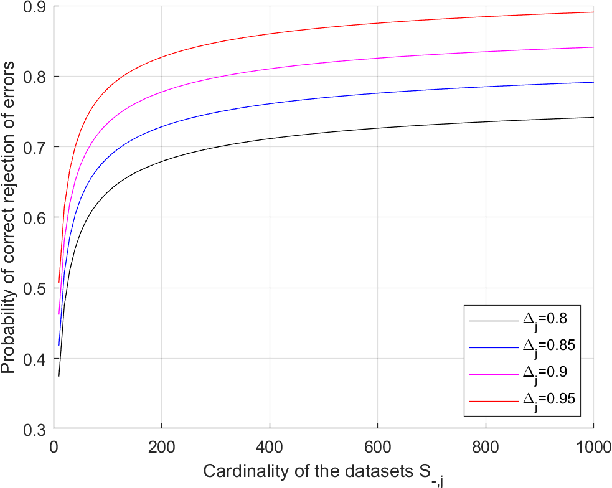

Abstract:We present a new methodology for handling AI errors by introducing weakly supervised AI error correctors with a priori performance guarantees. These AI correctors are auxiliary maps whose role is to moderate the decisions of some previously constructed underlying classifier by either approving or rejecting its decisions. The rejection of a decision can be used as a signal to suggest abstaining from making a decision. A key technical focus of the work is in providing performance guarantees for these new AI correctors through bounds on the probabilities of incorrect decisions. These bounds are distribution agnostic and do not rely on assumptions on the data dimension. Our empirical example illustrates how the framework can be applied to improve the performance of an image classifier in a challenging real-world task where training data are scarce.
An Informational Space Based Semantic Analysis for Scientific Texts
May 31, 2022



Abstract:One major problem in Natural Language Processing is the automatic analysis and representation of human language. Human language is ambiguous and deeper understanding of semantics and creating human-to-machine interaction have required an effort in creating the schemes for act of communication and building common-sense knowledge bases for the 'meaning' in texts. This paper introduces computational methods for semantic analysis and the quantifying the meaning of short scientific texts. Computational methods extracting semantic feature are used to analyse the relations between texts of messages and 'representations of situations' for a newly created large collection of scientific texts, Leicester Scientific Corpus. The representation of scientific-specific meaning is standardised by replacing the situation representations, rather than psychological properties, with the vectors of some attributes: a list of scientific subject categories that the text belongs to. First, this paper introduces 'Meaning Space' in which the informational representation of the meaning is extracted from the occurrence of the word in texts across the scientific categories, i.e., the meaning of a word is represented by a vector of Relative Information Gain about the subject categories. Then, the meaning space is statistically analysed for Leicester Scientific Dictionary-Core and we investigate 'Principal Components of the Meaning' to describe the adequate dimensions of the meaning. The research in this paper conducts the base for the geometric representation of the meaning of texts.
* 19 pages. arXiv admin note: substantial text overlap with arXiv:2009.08859, arXiv:2004.13717
Quasi-orthogonality and intrinsic dimensions as measures of learning and generalisation
Mar 30, 2022



Abstract:Finding best architectures of learning machines, such as deep neural networks, is a well-known technical and theoretical challenge. Recent work by Mellor et al (2021) showed that there may exist correlations between the accuracies of trained networks and the values of some easily computable measures defined on randomly initialised networks which may enable to search tens of thousands of neural architectures without training. Mellor et al used the Hamming distance evaluated over all ReLU neurons as such a measure. Motivated by these findings, in our work, we ask the question of the existence of other and perhaps more principled measures which could be used as determinants of success of a given neural architecture. In particular, we examine, if the dimensionality and quasi-orthogonality of neural networks' feature space could be correlated with the network's performance after training. We showed, using the setup as in Mellor et al, that dimensionality and quasi-orthogonality may jointly serve as network's performance discriminants. In addition to offering new opportunities to accelerate neural architecture search, our findings suggest important relationships between the networks' final performance and properties of their randomly initialised feature spaces: data dimension and quasi-orthogonality.
Scikit-dimension: a Python package for intrinsic dimension estimation
Sep 06, 2021



Abstract:Dealing with uncertainty in applications of machine learning to real-life data critically depends on the knowledge of intrinsic dimensionality (ID). A number of methods have been suggested for the purpose of estimating ID, but no standard package to easily apply them one by one or all at once has been implemented in Python. This technical note introduces \texttt{scikit-dimension}, an open-source Python package for intrinsic dimension estimation. \texttt{scikit-dimension} package provides a uniform implementation of most of the known ID estimators based on scikit-learn application programming interface to evaluate global and local intrinsic dimension, as well as generators of synthetic toy and benchmark datasets widespread in the literature. The package is developed with tools assessing the code quality, coverage, unit testing and continuous integration. We briefly describe the package and demonstrate its use in a large-scale (more than 500 datasets) benchmarking of methods for ID estimation in real-life and synthetic data. The source code is available from https://github.com/j-bac/scikit-dimension , the documentation is available from https://scikit-dimension.readthedocs.io .
Learning from scarce information: using synthetic data to classify Roman fine ware pottery
Jul 03, 2021



Abstract:In this article we consider a version of the challenging problem of learning from datasets whose size is too limited to allow generalisation beyond the training set. To address the challenge we propose to use a transfer learning approach whereby the model is first trained on a synthetic dataset replicating features of the original objects. In this study the objects were smartphone photographs of near-complete Roman terra sigillata pottery vessels from the collection of the Museum of London. Taking the replicated features from published profile drawings of pottery forms allowed the integration of expert knowledge into the process through our synthetic data generator. After this first initial training the model was fine-tuned with data from photographs of real vessels. We show, through exhaustive experiments across several popular deep learning architectures, different test priors, and considering the impact of the photograph viewpoint and excessive damage to the vessels, that the proposed hybrid approach enables the creation of classifiers with appropriate generalisation performance. This performance is significantly better than that of classifiers trained exclusively on the original data which shows the promise of the approach to alleviate the fundamental issue of learning from small datasets.
High-dimensional separability for one- and few-shot learning
Jun 28, 2021

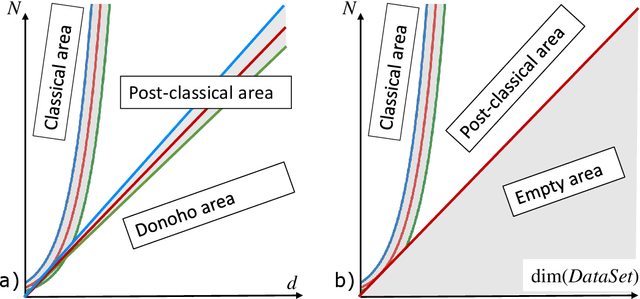
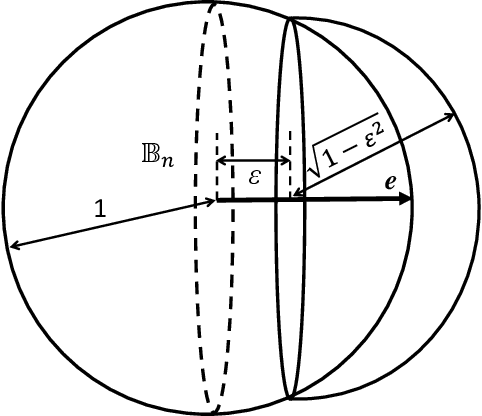
Abstract:This work is driven by a practical question, corrections of Artificial Intelligence (AI) errors. Systematic re-training of a large AI system is hardly possible. To solve this problem, special external devices, correctors, are developed. They should provide quick and non-iterative system fix without modification of a legacy AI system. A common universal part of the AI corrector is a classifier that should separate undesired and erroneous behavior from normal operation. Training of such classifiers is a grand challenge at the heart of the one- and few-shot learning methods. Effectiveness of one- and few-short methods is based on either significant dimensionality reductions or the blessing of dimensionality effects. Stochastic separability is a blessing of dimensionality phenomenon that allows one-and few-shot error correction: in high-dimensional datasets under broad assumptions each point can be separated from the rest of the set by simple and robust linear discriminant. The hierarchical structure of data universe is introduced where each data cluster has a granular internal structure, etc. New stochastic separation theorems for the data distributions with fine-grained structure are formulated and proved. Separation theorems in infinite-dimensional limits are proven under assumptions of compact embedding of patterns into data space. New multi-correctors of AI systems are presented and illustrated with examples of predicting errors and learning new classes of objects by a deep convolutional neural network.
Trajectories, bifurcations and pseudotime in large clinical datasets: applications to myocardial infarction and diabetes data
Jul 07, 2020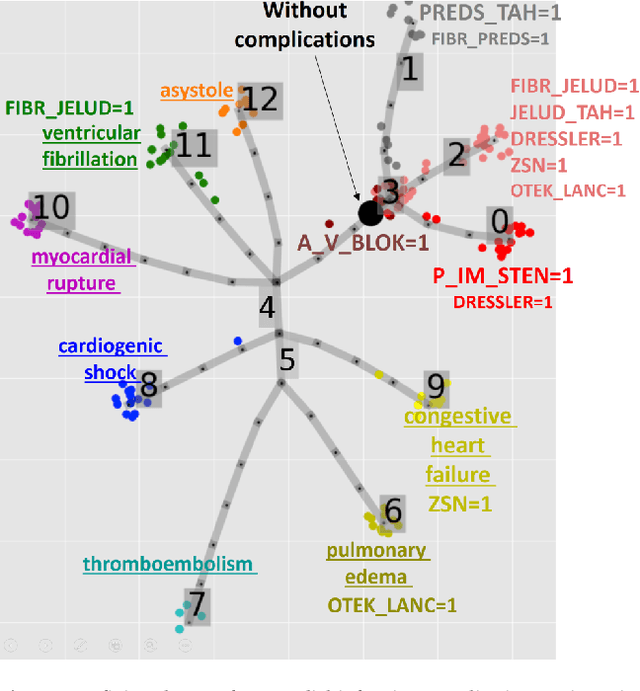
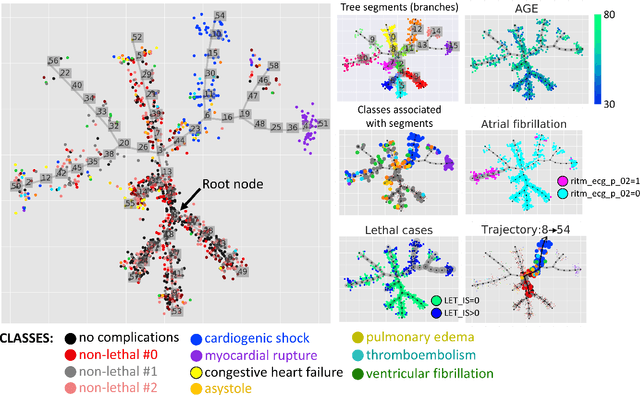
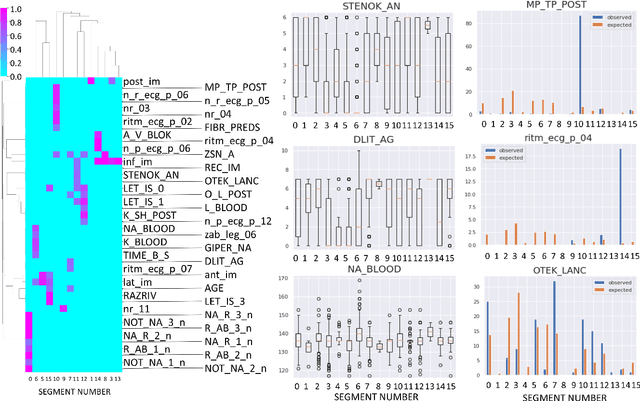
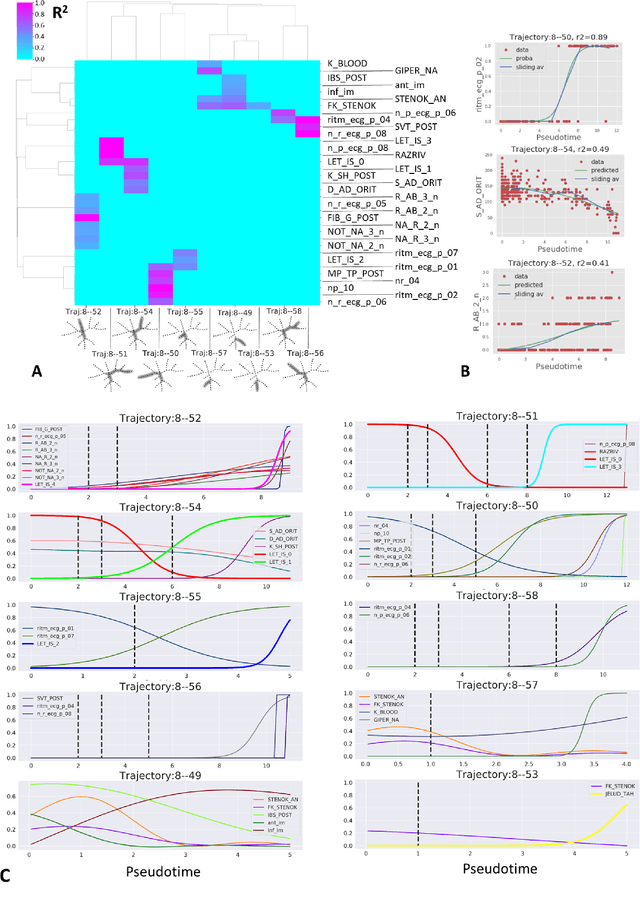
Abstract:Large observational clinical datasets become increasingly available for mining associations between various disease traits and administered therapy. These datasets can be considered as representations of the landscape of all possible disease conditions, in which a concrete pathology develops through a number of stereotypical routes, characterized by `points of no return' and `final states' (such as lethal or recovery states). Extracting this information directly from the data remains challenging, especially in the case of synchronic (with a short-term follow up) observations. Here we suggest a semi-supervised methodology for the analysis of large clinical datasets, characterized by mixed data types and missing values, through modeling the geometrical data structure as a bouquet of bifurcating clinical trajectories. The methodology is based on application of elastic principal graphs which can address simultaneously the tasks of dimensionality reduction, data visualization, clustering, feature selection and quantifying the geodesic distances (pseudotime) in partially ordered sequences of observations. The methodology allows positioning a patient on a particular clinical trajectory (pathological scenario) and characterizing the degree of progression along it with a qualitative estimate of the uncertainty of the prognosis. Overall, our pseudo-time quantification-based approach gives a possibility to apply the methods developed for dynamical disease phenotyping and illness trajectory analysis (diachronic data analysis) to synchronic observational data. We developed a tool $ClinTrajan$ for clinical trajectory analysis implemented in Python programming language. We test the methodology in two large publicly available datasets: myocardial infarction complications and readmission of diabetic patients data.
Fractional norms and quasinorms do not help to overcome the curse of dimensionality
Apr 29, 2020



Abstract:The curse of dimensionality causes the well-known and widely discussed problems for machine learning methods. There is a hypothesis that using of the Manhattan distance and even fractional quasinorms lp (for p less than 1) can help to overcome the curse of dimensionality in classification problems. In this study, we systematically test this hypothesis. We confirm that fractional quasinorms have a greater relative contrast or coefficient of variation than the Euclidean norm l2, but we also demonstrate that the distance concentration shows qualitatively the same behaviour for all tested norms and quasinorms and the difference between them decays as dimension tends to infinity. Estimation of classification quality for kNN based on different norms and quasinorms shows that a greater relative contrast does not mean better classifier performance and the worst performance for different databases was shown by different norms (quasinorms). A systematic comparison shows that the difference of the performance of kNN based on lp for p=2, 1, and 0.5 is statistically insignificant.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge