E. Kelly Buchanan
Terminal-Bench: Benchmarking Agents on Hard, Realistic Tasks in Command Line Interfaces
Jan 17, 2026Abstract:AI agents may soon become capable of autonomously completing valuable, long-horizon tasks in diverse domains. Current benchmarks either do not measure real-world tasks, or are not sufficiently difficult to meaningfully measure frontier models. To this end, we present Terminal-Bench 2.0: a carefully curated hard benchmark composed of 89 tasks in computer terminal environments inspired by problems from real workflows. Each task features a unique environment, human-written solution, and comprehensive tests for verification. We show that frontier models and agents score less than 65\% on the benchmark and conduct an error analysis to identify areas for model and agent improvement. We publish the dataset and evaluation harness to assist developers and researchers in future work at https://www.tbench.ai/ .
Brain-to-Text Benchmark '24: Lessons Learned
Dec 23, 2024

Abstract:Speech brain-computer interfaces aim to decipher what a person is trying to say from neural activity alone, restoring communication to people with paralysis who have lost the ability to speak intelligibly. The Brain-to-Text Benchmark '24 and associated competition was created to foster the advancement of decoding algorithms that convert neural activity to text. Here, we summarize the lessons learned from the competition ending on June 1, 2024 (the top 4 entrants also presented their experiences in a recorded webinar). The largest improvements in accuracy were achieved using an ensembling approach, where the output of multiple independent decoders was merged using a fine-tuned large language model (an approach used by all 3 top entrants). Performance gains were also found by improving how the baseline recurrent neural network (RNN) model was trained, including by optimizing learning rate scheduling and by using a diphone training objective. Improving upon the model architecture itself proved more difficult, however, with attempts to use deep state space models or transformers not yet appearing to offer a benefit over the RNN baseline. The benchmark will remain open indefinitely to support further work towards increasing the accuracy of brain-to-text algorithms.
Pathologies of Predictive Diversity in Deep Ensembles
Feb 03, 2023



Abstract:Classical results establish that ensembles of small models benefit when predictive diversity is encouraged, through bagging, boosting, and similar. Here we demonstrate that this intuition does not carry over to ensembles of deep neural networks used for classification, and in fact the opposite can be true. Unlike regression models or small (unconfident) classifiers, predictions from large (confident) neural networks concentrate in vertices of the probability simplex. Thus, decorrelating these points necessarily moves the ensemble prediction away from vertices, harming confidence and moving points across decision boundaries. Through large scale experiments, we demonstrate that diversity-encouraging regularizers hurt the performance of high-capacity deep ensembles used for classification. Even more surprisingly, discouraging predictive diversity can be beneficial. Together this work strongly suggests that the best strategy for deep ensembles is utilizing more accurate, but likely less diverse, component models.
Deep Ensembles Work, But Are They Necessary?
Feb 14, 2022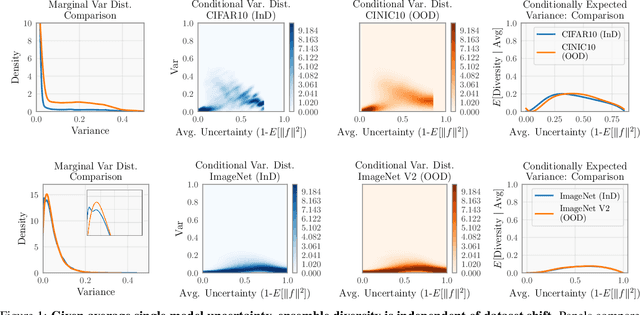
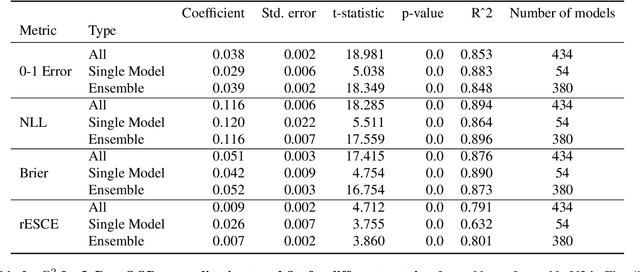
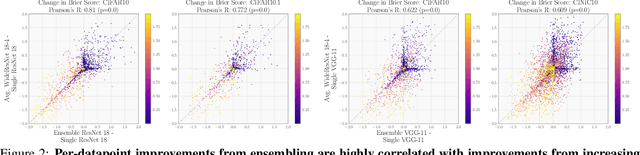
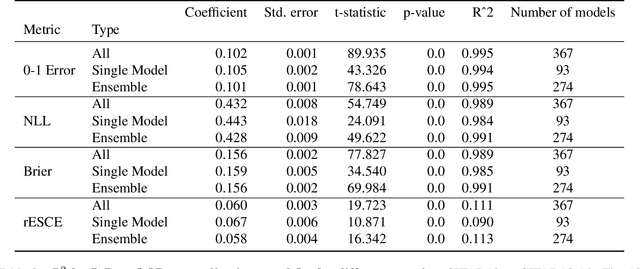
Abstract:Ensembling neural networks is an effective way to increase accuracy, and can often match the performance of larger models. This observation poses a natural question: given the choice between a deep ensemble and a single neural network with similar accuracy, is one preferable over the other? Recent work suggests that deep ensembles may offer benefits beyond predictive power: namely, uncertainty quantification and robustness to dataset shift. In this work, we demonstrate limitations to these purported benefits, and show that a single (but larger) neural network can replicate these qualities. First, we show that ensemble diversity, by any metric, does not meaningfully contribute to an ensemble's ability to detect out-of-distribution (OOD) data, and that one can estimate ensemble diversity by measuring the relative improvement of a single larger model. Second, we show that the OOD performance afforded by ensembles is strongly determined by their in-distribution (InD) performance, and -- in this sense -- is not indicative of any "effective robustness". While deep ensembles are a practical way to achieve performance improvement (in agreement with prior work), our results show that they may be a tool of convenience rather than a fundamentally better model class.
Penalized matrix decomposition for denoising, compression, and improved demixing of functional imaging data
Jul 17, 2018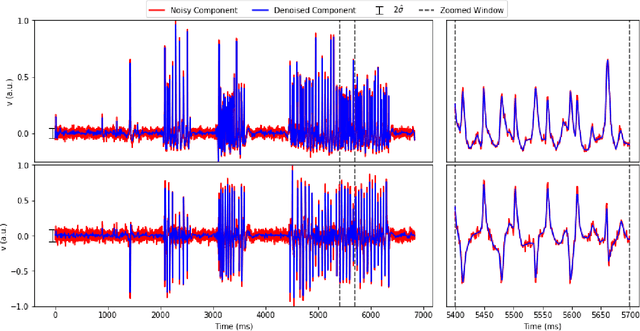
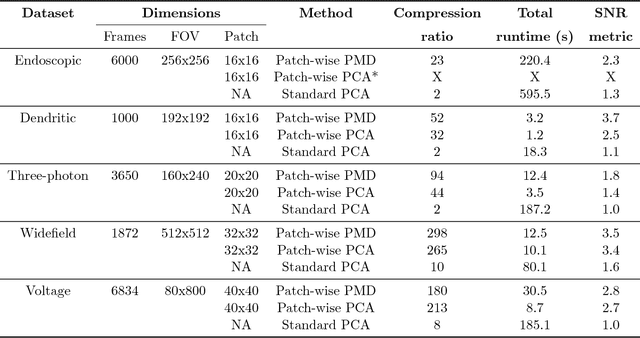

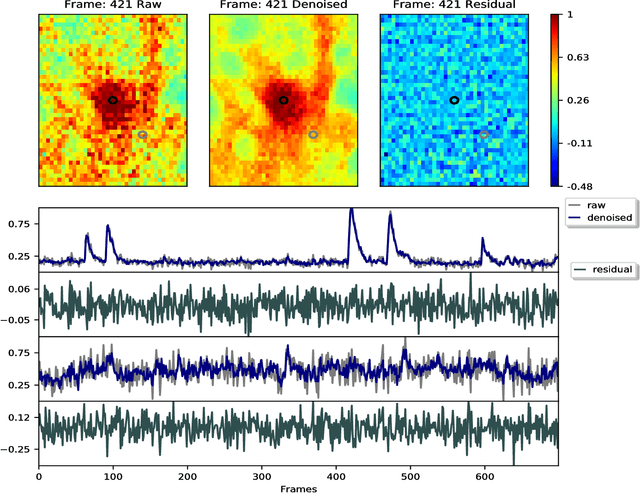
Abstract:Calcium imaging has revolutionized systems neuroscience, providing the ability to image large neural populations with single-cell resolution. The resulting datasets are quite large, which has presented a barrier to routine open sharing of this data, slowing progress in reproducible research. State of the art methods for analyzing this data are based on non-negative matrix factorization (NMF); these approaches solve a non-convex optimization problem, and are effective when good initializations are available, but can break down in low-SNR settings where common initialization approaches fail. Here we introduce an approach to compressing and denoising functional imaging data. The method is based on a spatially-localized penalized matrix decomposition (PMD) of the data to separate (low-dimensional) signal from (temporally-uncorrelated) noise. This approach can be applied in parallel on local spatial patches and is therefore highly scalable, does not impose non-negativity constraints or require stringent identifiability assumptions (leading to significantly more robust results compared to NMF), and estimates all parameters directly from the data, so no hand-tuning is required. We have applied the method to a wide range of functional imaging data (including one-photon, two-photon, three-photon, widefield, somatic, axonal, dendritic, calcium, and voltage imaging datasets): in all cases, we observe ~2-4x increases in SNR and compression rates of 20-300x with minimal visible loss of signal, with no adjustment of hyperparameters; this in turn facilitates the process of demixing the observed activity into contributions from individual neurons. We focus on two challenging applications: dendritic calcium imaging data and voltage imaging data in the context of optogenetic stimulation. In both cases, we show that our new approach leads to faster and much more robust extraction of activity from the data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge