Douglas Weber
Interpretation of Intracardiac Electrograms Through Textual Representations
Feb 02, 2024



Abstract:Understanding the irregular electrical activity of atrial fibrillation (AFib) has been a key challenge in electrocardiography. For serious cases of AFib, catheter ablations are performed to collect intracardiac electrograms (EGMs). EGMs offer intricately detailed and localized electrical activity of the heart and are an ideal modality for interpretable cardiac studies. Recent advancements in artificial intelligence (AI) has allowed some works to utilize deep learning frameworks to interpret EGMs during AFib. Additionally, language models (LMs) have shown exceptional performance in being able to generalize to unseen domains, especially in healthcare. In this study, we are the first to leverage pretrained LMs for finetuning of EGM interpolation and AFib classification via masked language modeling. We formulate the EGM as a textual sequence and present competitive performances on AFib classification compared against other representations. Lastly, we provide a comprehensive interpretability study to provide a multi-perspective intuition of the model's behavior, which could greatly benefit the clinical use.
High-density Electromyography for Effective Gesture-based Control of Physically Assistive Mobile Manipulators
Dec 12, 2023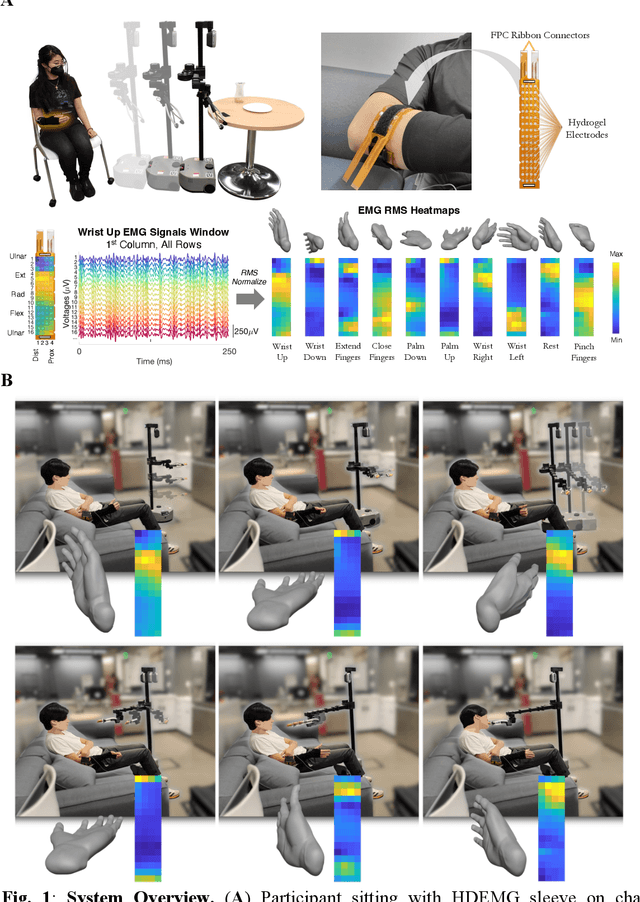
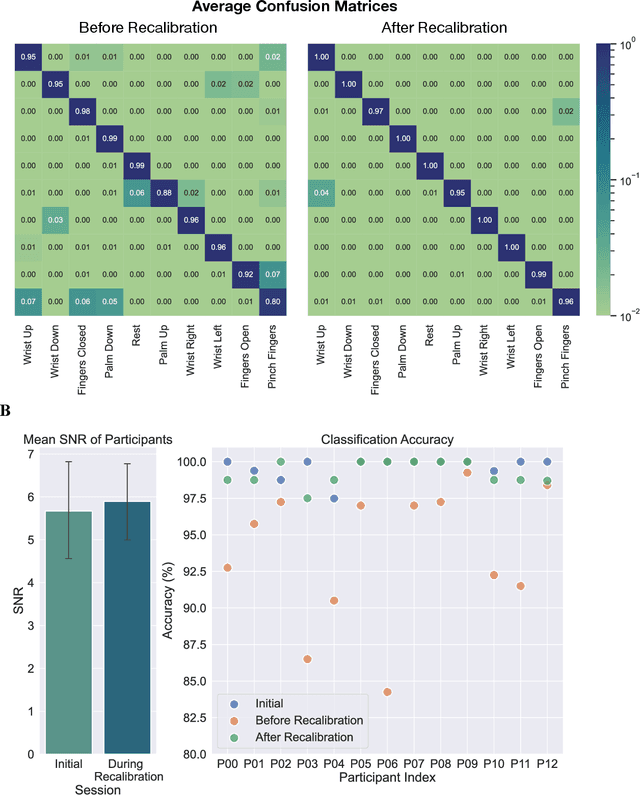
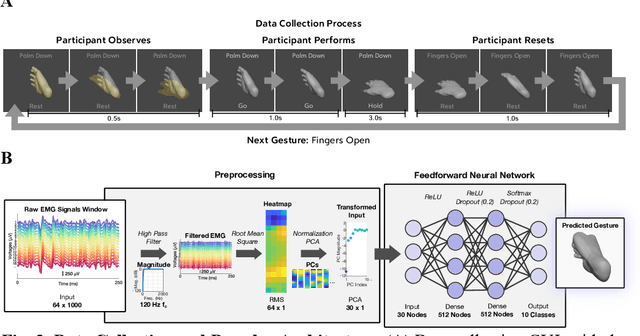
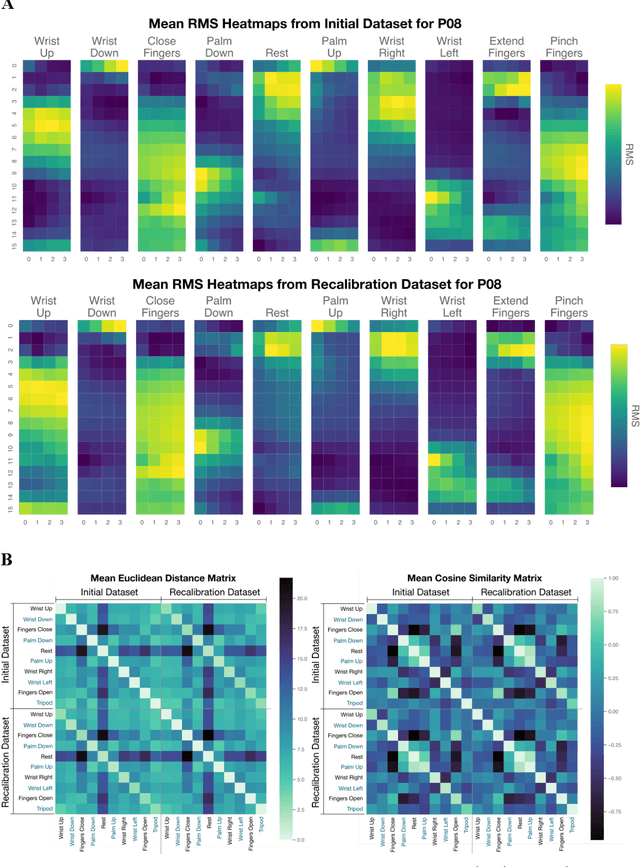
Abstract:Injury to the cervical spinal cord can cause quadriplegia, impairing muscle function in all four limbs. People with impaired hand function and mobility encounter significant difficulties in carrying out essential self-care and household tasks. Despite the impairment of their neural drive, their volitional myoelectric activity is often partially preserved. High-density electromyography (HDEMG) can detect this myoelectric activity, which can serve as control inputs to assistive devices. Previous HDEMG-controlled robotic interfaces have primarily been limited to controlling table-mounted robot arms. These have constrained reach capabilities. Instead, the ability to control mobile manipulators, which have no such workspace constraints, could allow individuals with quadriplegia to perform a greater variety of assistive tasks, thus restoring independence and reducing caregiver workload. In this study, we introduce a non-invasive wearable HDEMG interface with real-time myoelectric hand gesture recognition, enabling both coarse and fine control over the intricate mobility and manipulation functionalities of an 8 degree-of-freedom mobile manipulator. Our evaluation, involving 13 participants engaging in challenging self-care and household activities, demonstrates the potential of our wearable HDEMG system to profoundly enhance user independence by enabling non-invasive control of a mobile manipulator.
Multimodal Representation Learning of Cardiovascular Magnetic Resonance Imaging
Apr 16, 2023Abstract:Self-supervised learning is crucial for clinical imaging applications, given the lack of explicit labels in healthcare. However, conventional approaches that rely on precise vision-language alignment are not always feasible in complex clinical imaging modalities, such as cardiac magnetic resonance (CMR). CMR provides a comprehensive visualization of cardiac anatomy, physiology, and microstructure, making it challenging to interpret. Additionally, CMR reports require synthesizing information from sequences of images and different views, resulting in potentially weak alignment between the study and diagnosis report pair. To overcome these challenges, we propose \textbf{CMRformer}, a multimodal learning framework to jointly learn sequences of CMR images and associated cardiologist's reports. Moreover, one of the major obstacles to improving CMR study is the lack of large, publicly available datasets. To bridge this gap, we collected a large \textbf{CMR dataset}, which consists of 13,787 studies from clinical cases. By utilizing our proposed CMRformer and our collected dataset, we achieved remarkable performance in real-world clinical tasks, such as CMR image retrieval and diagnosis report retrieval. Furthermore, the learned representations are evaluated to be practically helpful for downstream applications, such as disease classification. Our work could potentially expedite progress in the CMR study and lead to more accurate and effective diagnosis and treatment.
Converting ECG Signals to Images for Efficient Image-text Retrieval via Encoding
Apr 13, 2023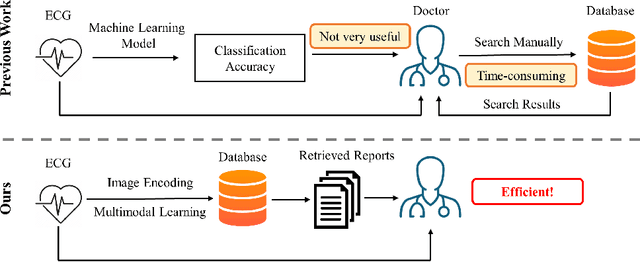
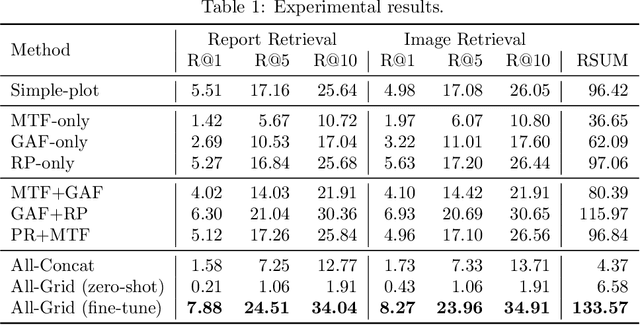
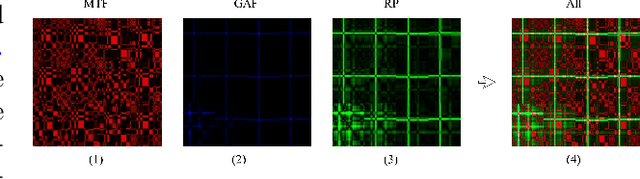
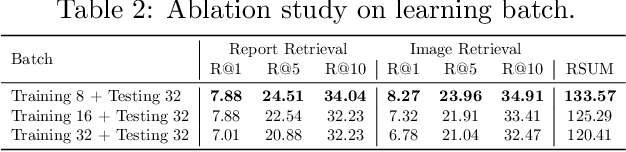
Abstract:Automated interpretation of electrocardiograms (ECG) has garnered significant attention with the advancements in machine learning methodologies. Despite the growing interest in automated ECG interpretation using machine learning, most current studies focus solely on classification or regression tasks and overlook a crucial aspect of clinical cardio-disease diagnosis: the diagnostic report generated by experienced human clinicians. In this paper, we introduce a novel approach to ECG interpretation, leveraging recent breakthroughs in Large Language Models (LLMs) and Vision-Transformer (ViT) models. Rather than treating ECG diagnosis as a classification or regression task, we propose an alternative method of automatically identifying the most similar clinical cases based on the input ECG data. Also, since interpreting ECG as images are more affordable and accessible, we process ECG as encoded images and adopt a vision-language learning paradigm to jointly learn vision-language alignment between encoded ECG images and ECG diagnosis reports. Encoding ECG into images can result in an efficient ECG retrieval system, which will be highly practical and useful in clinical applications. More importantly, our findings could serve as a crucial resource for providing diagnostic services in regions where only paper-printed ECG images are accessible due to past underdevelopment.
Transfer Knowledge from Natural Language to Electrocardiography: Can We Detect Cardiovascular Disease Through Language Models?
Jan 21, 2023



Abstract:Recent advancements in Large Language Models (LLMs) have drawn increasing attention since the learned embeddings pretrained on large-scale datasets have shown powerful ability in various downstream applications. However, whether the learned knowledge by LLMs can be transferred to clinical cardiology remains unknown. In this work, we aim to bridge this gap by transferring the knowledge of LLMs to clinical Electrocardiography (ECG). We propose an approach for cardiovascular disease diagnosis and automatic ECG diagnosis report generation. We also introduce an additional loss function by Optimal Transport (OT) to align the distribution between ECG and language embedding. The learned embeddings are evaluated on two downstream tasks: (1) automatic ECG diagnosis report generation, and (2) zero-shot cardiovascular disease detection. Our approach is able to generate high-quality cardiac diagnosis reports and also achieves competitive zero-shot classification performance even compared with supervised baselines, which proves the feasibility of transferring knowledge from LLMs to the cardiac domain.
An Empirical Exploration of Cross-domain Alignment between Language and Electroencephalogram
Aug 21, 2022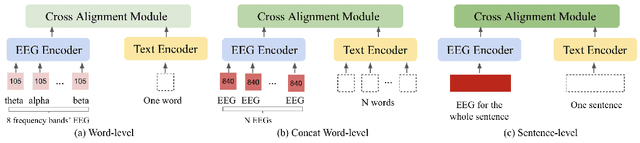
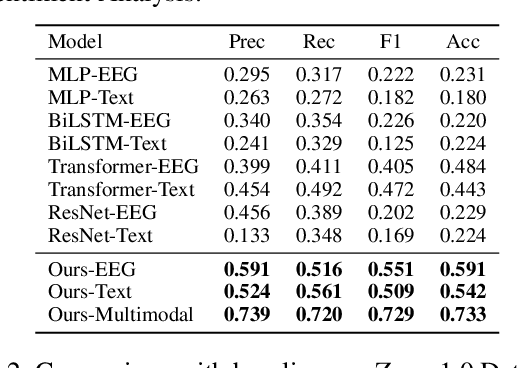
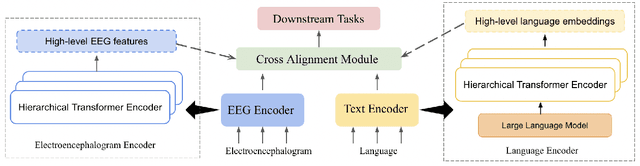
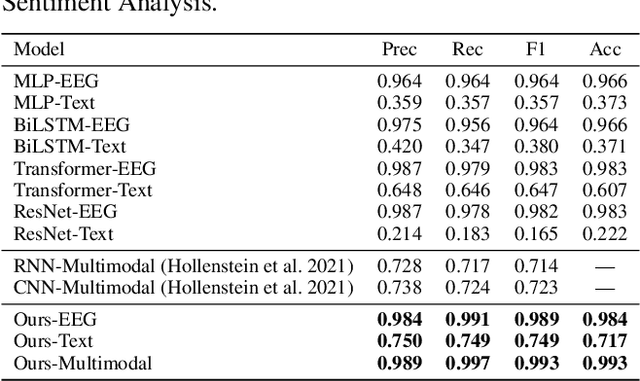
Abstract:Electroencephalography (EEG) and language have been widely explored independently for many downstream tasks (e.g., sentiment analysis, relation detection, etc.). Multimodal approaches that study both domains have not been well explored, even though in recent years, multimodal learning has been seen to be more powerful than its unimodal counterparts. In this study, we want to explore the relationship and dependency between EEG and language, i.e., how one domain reflects and represents the other. To study the relationship at the representation level, we introduced MTAM, a MultimodalTransformer Alignment Model, to observe coordinated representations between the two modalities, and thus employ the transformed representations for downstream applications. We used various relationship alignment-seeking techniques, such as Canonical Correlation Analysis and Wasserstein Distance, as loss functions to transfigure low-level language and EEG features to high-level transformed features. On downstream applications, sentiment analysis and relation detection, we achieved new state-of-the-art results on two datasets, ZuCo and K-EmoCon. Our method achieved an F1-score improvement of 16.5% on sentiment analysis for K-EmoCon, 27% on sentiment analysis of ZuCo, and 31.1% on relation detection of ZuCo. In addition, we provide interpretations of the performance improvement by: (1) visualizing the original feature distribution and the transformed feature distribution, showing the effectiveness of the alignment module for discovering and encoding the relationship between EEG and language; (2) visualizing word-level and sentence-level EEG-language alignment weights, showing the influence of different language semantics as well as EEG frequency features; and (3) visualizing brain topographical maps to provide an intuitive demonstration of the connectivity of EEG and language response in the brain regions.
GeoECG: Data Augmentation via Wasserstein Geodesic Perturbation for Robust Electrocardiogram Prediction
Aug 10, 2022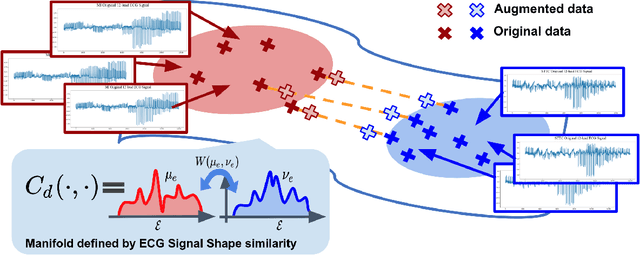
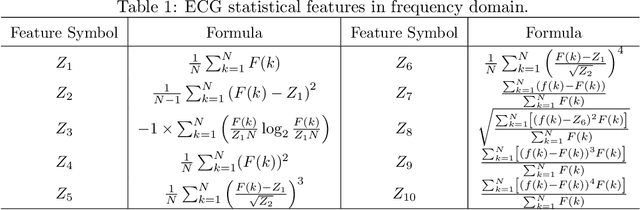
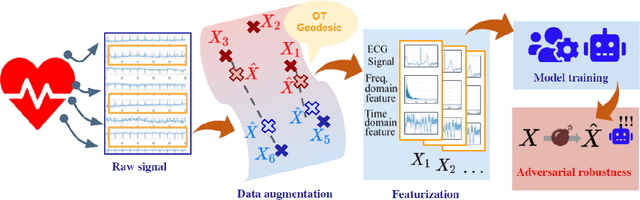
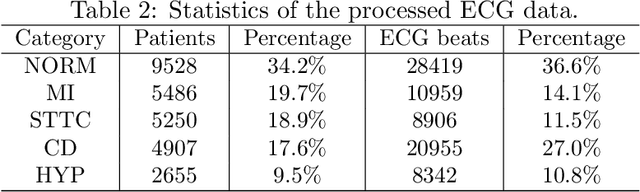
Abstract:There has been an increased interest in applying deep neural networks to automatically interpret and analyze the 12-lead electrocardiogram (ECG). The current paradigms with machine learning methods are often limited by the amount of labeled data. This phenomenon is particularly problematic for clinically-relevant data, where labeling at scale can be time-consuming and costly in terms of the specialized expertise and human effort required. Moreover, deep learning classifiers may be vulnerable to adversarial examples and perturbations, which could have catastrophic consequences, for example, when applied in the context of medical treatment, clinical trials, or insurance claims. In this paper, we propose a physiologically-inspired data augmentation method to improve performance and increase the robustness of heart disease detection based on ECG signals. We obtain augmented samples by perturbing the data distribution towards other classes along the geodesic in Wasserstein space. To better utilize domain-specific knowledge, we design a ground metric that recognizes the difference between ECG signals based on physiologically determined features. Learning from 12-lead ECG signals, our model is able to distinguish five categories of cardiac conditions. Our results demonstrate improvements in accuracy and robustness, reflecting the effectiveness of our data augmentation method.
* 26 pages, Figure 13, Machine Learning for Healthcare 2022
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge