Daniel Daza
Explaining Graph Neural Networks for Node Similarity on Graphs
Jul 10, 2024Abstract:Similarity search is a fundamental task for exploiting information in various applications dealing with graph data, such as citation networks or knowledge graphs. While this task has been intensively approached from heuristics to graph embeddings and graph neural networks (GNNs), providing explanations for similarity has received less attention. In this work we are concerned with explainable similarity search over graphs, by investigating how GNN-based methods for computing node similarities can be augmented with explanations. Specifically, we evaluate the performance of two prominent approaches towards explanations in GNNs, based on the concepts of mutual information (MI), and gradient-based explanations (GB). We discuss their suitability and empirically validate the properties of their explanations over different popular graph benchmarks. We find that unlike MI explanations, gradient-based explanations have three desirable properties. First, they are actionable: selecting inputs depending on them results in predictable changes in similarity scores. Second, they are consistent: the effect of selecting certain inputs overlaps very little with the effect of discarding them. Third, they can be pruned significantly to obtain sparse explanations that retain the effect on similarity scores.
GRAPES: Learning to Sample Graphs for Scalable Graph Neural Networks
Oct 05, 2023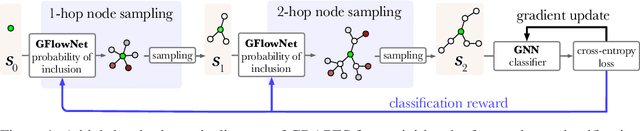
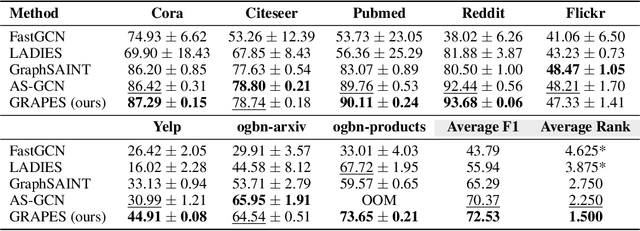
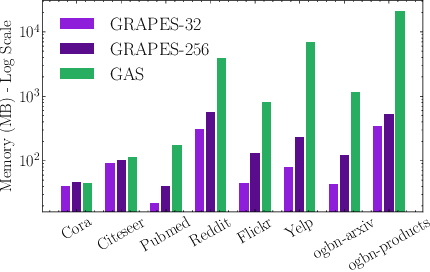
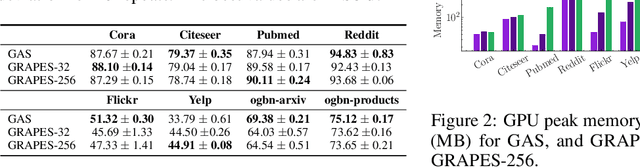
Abstract:Graph neural networks (GNNs) learn the representation of nodes in a graph by aggregating the neighborhood information in various ways. As these networks grow in depth, their receptive field grows exponentially due to the increase in neighborhood sizes, resulting in high memory costs. Graph sampling solves memory issues in GNNs by sampling a small ratio of the nodes in the graph. This way, GNNs can scale to much larger graphs. Most sampling methods focus on fixed sampling heuristics, which may not generalize to different structures or tasks. We introduce GRAPES, an adaptive graph sampling method that learns to identify sets of influential nodes for training a GNN classifier. GRAPES uses a GFlowNet to learn node sampling probabilities given the classification objectives. We evaluate GRAPES across several small- and large-scale graph benchmarks and demonstrate its effectiveness in accuracy and scalability. In contrast to existing sampling methods, GRAPES maintains high accuracy even with small sample sizes and, therefore, can scale to very large graphs. Our code is publicly available at https://github.com/dfdazac/grapes.
Approximate Answering of Graph Queries
Aug 12, 2023Abstract:Knowledge graphs (KGs) are inherently incomplete because of incomplete world knowledge and bias in what is the input to the KG. Additionally, world knowledge constantly expands and evolves, making existing facts deprecated or introducing new ones. However, we would still want to be able to answer queries as if the graph were complete. In this chapter, we will give an overview of several methods which have been proposed to answer queries in such a setting. We will first provide an overview of the different query types which can be supported by these methods and datasets typically used for evaluation, as well as an insight into their limitations. Then, we give an overview of the different approaches and describe them in terms of expressiveness, supported graph types, and inference capabilities.
Harnessing the Web and Knowledge Graphs for Automated Impact Investing Scoring
Aug 04, 2023



Abstract:The Sustainable Development Goals (SDGs) were introduced by the United Nations in order to encourage policies and activities that help guarantee human prosperity and sustainability. SDG frameworks produced in the finance industry are designed to provide scores that indicate how well a company aligns with each of the 17 SDGs. This scoring enables a consistent assessment of investments that have the potential of building an inclusive and sustainable economy. As a result of the high quality and reliability required by such frameworks, the process of creating and maintaining them is time-consuming and requires extensive domain expertise. In this work, we describe a data-driven system that seeks to automate the process of creating an SDG framework. First, we propose a novel method for collecting and filtering a dataset of texts from different web sources and a knowledge graph relevant to a set of companies. We then implement and deploy classifiers trained with this data for predicting scores of alignment with SDGs for a given company. Our results indicate that our best performing model can accurately predict SDG scores with a micro average F1 score of 0.89, demonstrating the effectiveness of the proposed solution. We further describe how the integration of the models for its use by humans can be facilitated by providing explanations in the form of data relevant to a predicted score. We find that our proposed solution enables access to a large amount of information that analysts would normally not be able to process, resulting in an accurate prediction of SDG scores at a fraction of the cost.
BioBLP: A Modular Framework for Learning on Multimodal Biomedical Knowledge Graphs
Jun 06, 2023Abstract:Knowledge graphs (KGs) are an important tool for representing complex relationships between entities in the biomedical domain. Several methods have been proposed for learning embeddings that can be used to predict new links in such graphs. Some methods ignore valuable attribute data associated with entities in biomedical KGs, such as protein sequences, or molecular graphs. Other works incorporate such data, but assume that entities can be represented with the same data modality. This is not always the case for biomedical KGs, where entities exhibit heterogeneous modalities that are central to their representation in the subject domain. We propose a modular framework for learning embeddings in KGs with entity attributes, that allows encoding attribute data of different modalities while also supporting entities with missing attributes. We additionally propose an efficient pretraining strategy for reducing the required training runtime. We train models using a biomedical KG containing approximately 2 million triples, and evaluate the performance of the resulting entity embeddings on the tasks of link prediction, and drug-protein interaction prediction, comparing against methods that do not take attribute data into account. In the standard link prediction evaluation, the proposed method results in competitive, yet lower performance than baselines that do not use attribute data. When evaluated in the task of drug-protein interaction prediction, the method compares favorably with the baselines. We find settings involving low degree entities, which make up for a substantial amount of the set of entities in the KG, where our method outperforms the baselines. Our proposed pretraining strategy yields significantly higher performance while reducing the required training runtime. Our implementation is available at https://github.com/elsevier-AI-Lab/BioBLP .
Approximate Knowledge Graph Query Answering: From Ranking to Binary Classification
Feb 22, 2021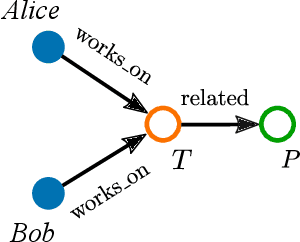
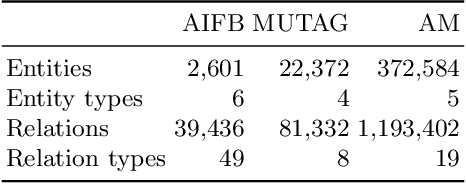
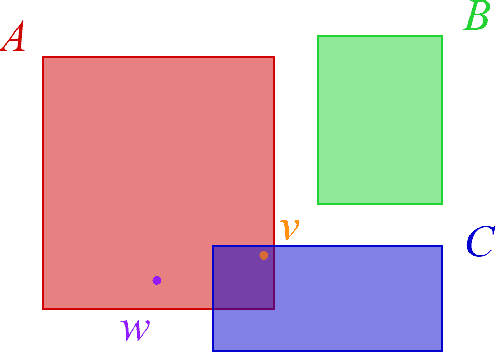
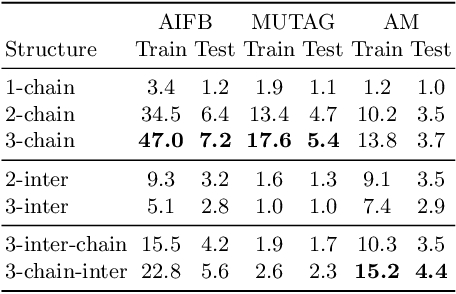
Abstract:Large, heterogeneous datasets are characterized by missing or even erroneous information. This is more evident when they are the product of community effort or automatic fact extraction methods from external sources, such as text. A special case of the aforementioned phenomenon can be seen in knowledge graphs, where this mostly appears in the form of missing or incorrect edges and nodes. Structured querying on such incomplete graphs will result in incomplete sets of answers, even if the correct entities exist in the graph, since one or more edges needed to match the pattern are missing. To overcome this problem, several algorithms for approximate structured query answering have been proposed. Inspired by modern Information Retrieval metrics, these algorithms produce a ranking of all entities in the graph, and their performance is further evaluated based on how high in this ranking the correct answers appear. In this work we take a critical look at this way of evaluation. We argue that performing a ranking-based evaluation is not sufficient to assess methods for complex query answering. To solve this, we introduce Message Passing Query Boxes (MPQB), which takes binary classification metrics back into use and shows the effect this has on the recently proposed query embedding method MPQE.
Complex Query Answering with Neural Link Predictors
Nov 06, 2020



Abstract:Neural link predictors are immensely useful for identifying missing edges in large scale Knowledge Graphs. However, it is still not clear how to use these models for answering more complex queries that arise in a number of domains, such as queries using logical conjunctions, disjunctions, and existential quantifiers, while accounting for missing edges. In this work, we propose a framework for efficiently answering complex queries on incomplete Knowledge Graphs. We translate each query into an end-to-end differentiable objective, where the truth value of each atom is computed by a pre-trained neural link predictor. We then analyse two solutions to the optimisation problem, including gradient-based and combinatorial search. In our experiments, the proposed approach produces more accurate results than state-of-the-art methods -- black-box neural models trained on millions of generated queries -- without the need of training on a large and diverse set of complex queries. Using orders of magnitude less training data, we obtain relative improvements ranging from 8% up to 40% in Hits@3 across different knowledge graphs containing factual information. Finally, we demonstrate that it is possible to explain the outcome of our model in terms of the intermediate solutions identified for each of the complex query atoms.
Inductive Entity Representations from Text via Link Prediction
Oct 07, 2020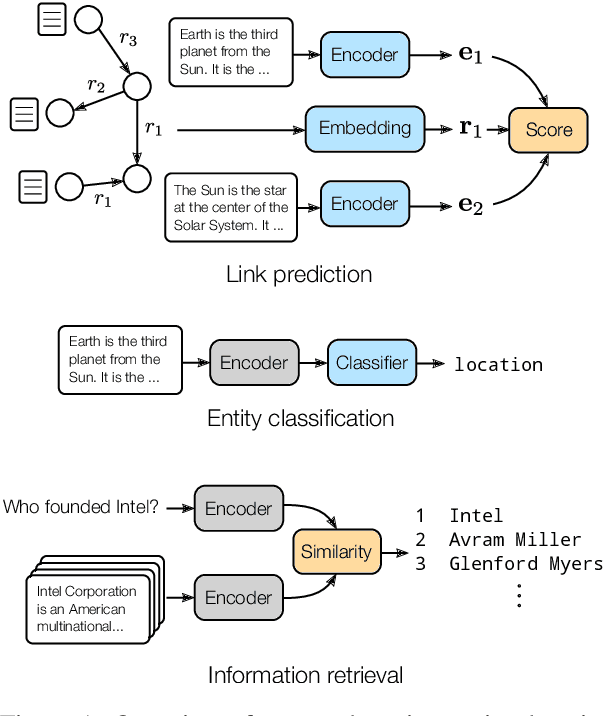
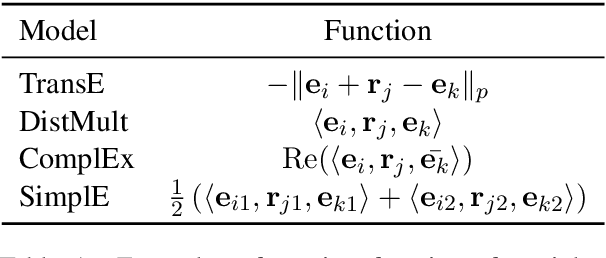
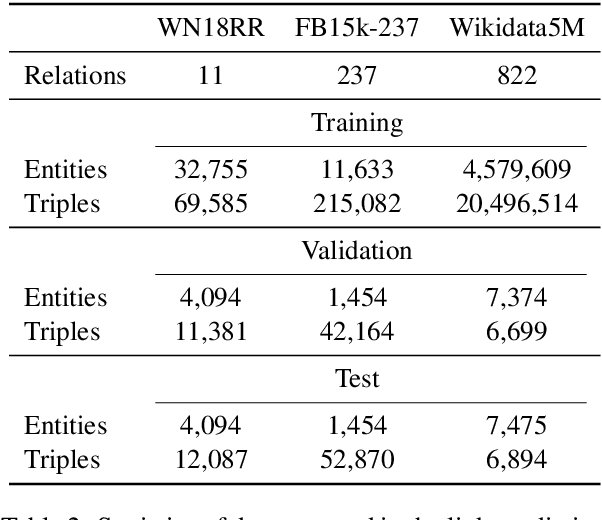
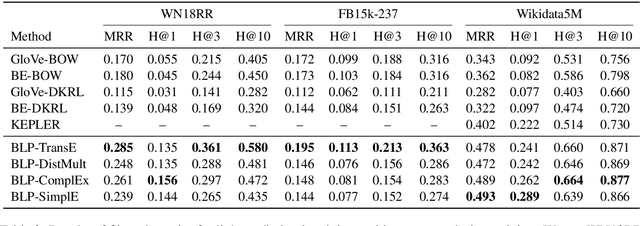
Abstract:We present a method for learning representations of entities, that uses a Transformer-based architecture as an entity encoder, and link prediction training on a knowledge graph with textual entity descriptions. We demonstrate that our approach can be applied effectively for link prediction in different inductive settings involving entities not seen during training, outperforming related state-of-the-art methods (22% MRR improvement on average). We provide evidence that the learned representations transfer to other tasks that do not require fine-tuning the entity encoder. In an entity classification task we obtain an average improvement of 16% accuracy compared with baselines that also employ pre-trained models. For an information retrieval task, significant improvements of up to 8.8% in NDCG@10 were obtained for natural language queries.
Message Passing for Query Answering over Knowledge Graphs
Feb 06, 2020



Abstract:Logic-based systems for query answering over knowledge graphs return only answers that rely on information explicitly represented in the graph. To improve recall, recent works have proposed the use of embeddings to predict additional information like missing links, or labels. These embeddings enable scoring entities in the graph as the answer a query, without being fully dependent on the graph structure. In its simplest case, answering a query in such a setting requires predicting a link between two entities. However, link prediction is not sufficient to address complex queries that involve multiple entities and variables. To solve this task, we propose to apply a message passing mechanism to a graph representation of the query, where nodes correspond to variables and entities. This results in an embedding of the query, such that answering entities are close to it in the embedding space. The general formulation of our method allows it to encode a more diverse set of query types in comparison to previous work. We evaluate our method by answering queries that rely on edges not seen during training, obtaining competitive performance. In contrast with previous work, we show that our method can generalize from training for the single-hop, link prediction task, to answering queries with more complex structures. A qualitative analysis reveals that the learned embeddings successfully capture the notion of different entity types.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge