Camilo Thorne
Learning Section Weights for Multi-Label Document Classification
Nov 26, 2023Abstract:Multi-label document classification is a traditional task in NLP. Compared to single-label classification, each document can be assigned multiple classes. This problem is crucially important in various domains, such as tagging scientific articles. Documents are often structured into several sections such as abstract and title. Current approaches treat different sections equally for multi-label classification. We argue that this is not a realistic assumption, leading to sub-optimal results. Instead, we propose a new method called Learning Section Weights (LSW), leveraging the contribution of each distinct section for multi-label classification. Via multiple feed-forward layers, LSW learns to assign weights to each section of, and incorporate the weights in the prediction. We demonstrate our approach on scientific articles. Experimental results on public (arXiv) and private (Elsevier) datasets confirm the superiority of LSW, compared to state-of-the-art multi-label document classification methods. In particular, LSW achieves a 1.3% improvement in terms of macro averaged F1-score while it achieves 1.3% in terms of macro averaged recall on the publicly available arXiv dataset.
One Strike, You're Out: Detecting Markush Structures in Low Signal-to-Noise Ratio Images
Nov 24, 2023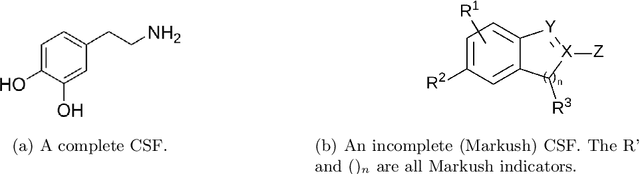
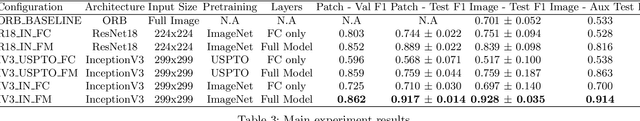
Abstract:Modern research increasingly relies on automated methods to assist researchers. An example of this is Optical Chemical Structure Recognition (OCSR), which aids chemists in retrieving information about chemicals from large amounts of documents. Markush structures are chemical structures that cannot be parsed correctly by OCSR and cause errors. The focus of this research was to propose and test a novel method for classifying Markush structures. Within this method, a comparison was made between fixed-feature extraction and end-to-end learning (CNN). The end-to-end method performed significantly better than the fixed-feature method, achieving 0.928 (0.035 SD) Macro F1 compared to the fixed-feature method's 0.701 (0.052 SD). Because of the nature of the experiment, these figures are a lower bound and can be improved further. These results suggest that Markush structures can be filtered out effectively and accurately using the proposed method. When implemented into OCSR pipelines, this method can improve their performance and use to other researchers.
Stress Testing BERT Anaphora Resolution Models for Reaction Extraction in Chemical Patents
Jun 23, 2023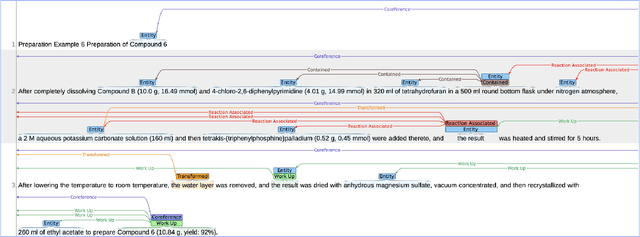
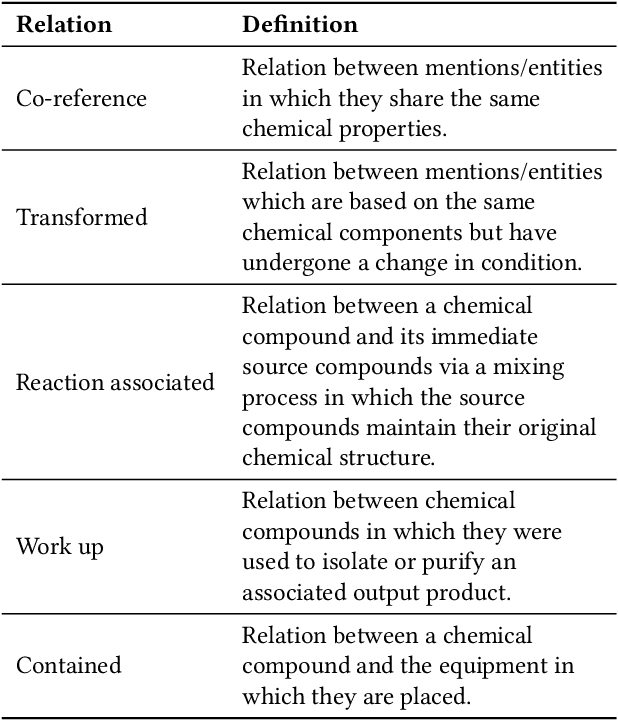
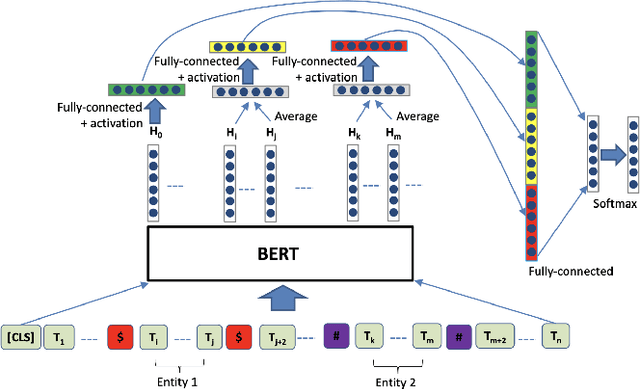
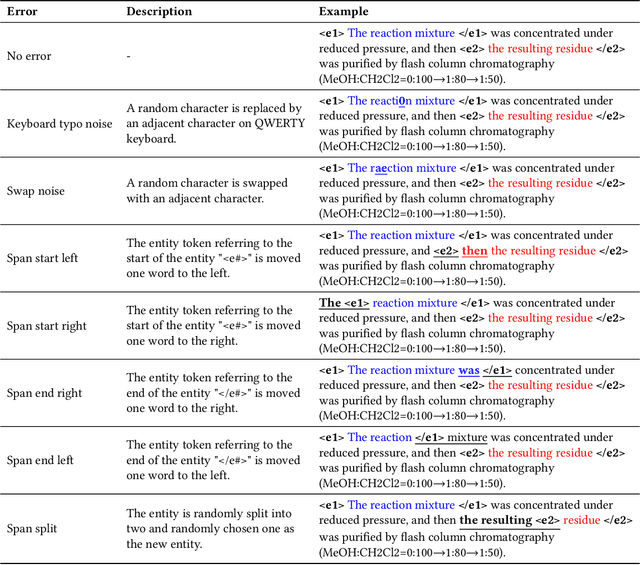
Abstract:The high volume of published chemical patents and the importance of a timely acquisition of their information gives rise to automating information extraction from chemical patents. Anaphora resolution is an important component of comprehensive information extraction, and is critical for extracting reactions. In chemical patents, there are five anaphoric relations of interest: co-reference, transformed, reaction associated, work up, and contained. Our goal is to investigate how the performance of anaphora resolution models for reaction texts in chemical patents differs in a noise-free and noisy environment and to what extent we can improve the robustness against noise of the model.
Stress Test Evaluation of Biomedical Word Embeddings
Jul 24, 2021
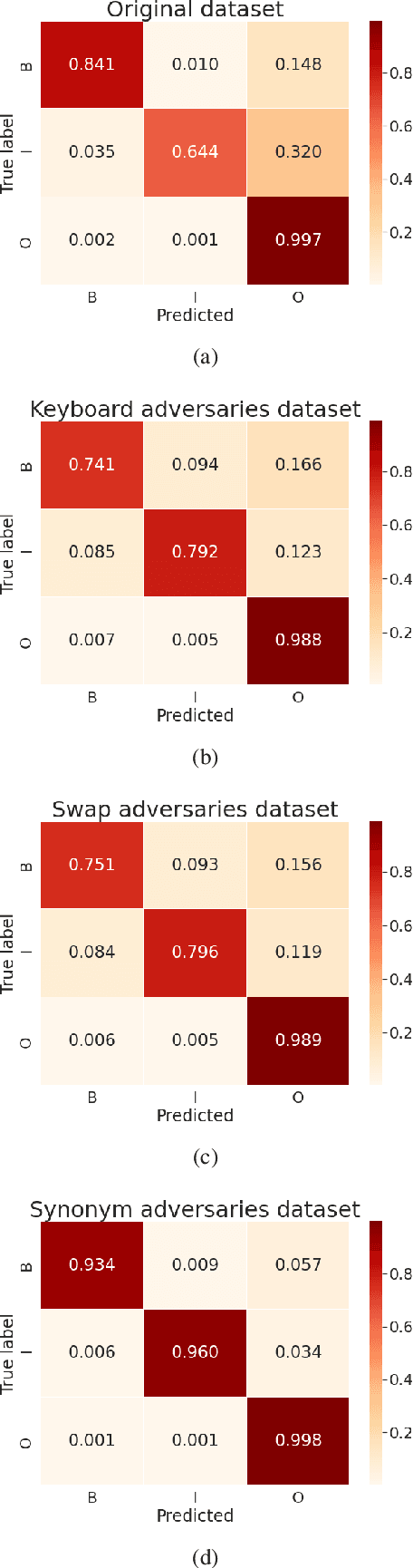
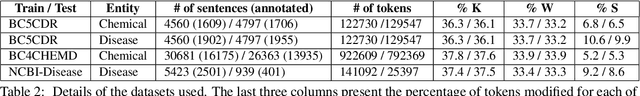
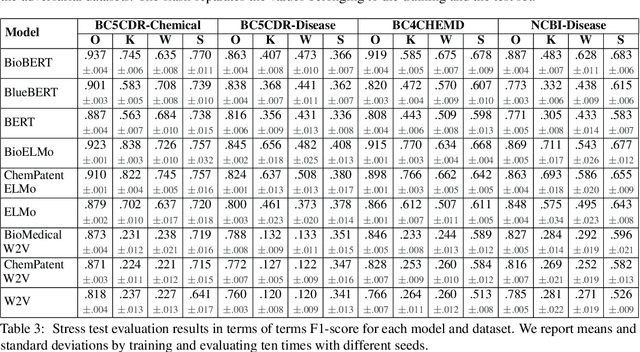
Abstract:The success of pretrained word embeddings has motivated their use in the biomedical domain, with contextualized embeddings yielding remarkable results in several biomedical NLP tasks. However, there is a lack of research on quantifying their behavior under severe "stress" scenarios. In this work, we systematically evaluate three language models with adversarial examples -- automatically constructed tests that allow us to examine how robust the models are. We propose two types of stress scenarios focused on the biomedical named entity recognition (NER) task, one inspired by spelling errors and another based on the use of synonyms for medical terms. Our experiments with three benchmarks show that the performance of the original models decreases considerably, in addition to revealing their weaknesses and strengths. Finally, we show that adversarial training causes the models to improve their robustness and even to exceed the original performance in some cases.
Disease Normalization with Graph Embeddings
Oct 24, 2020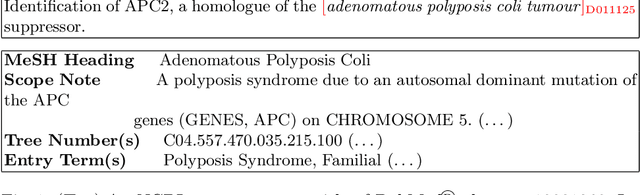
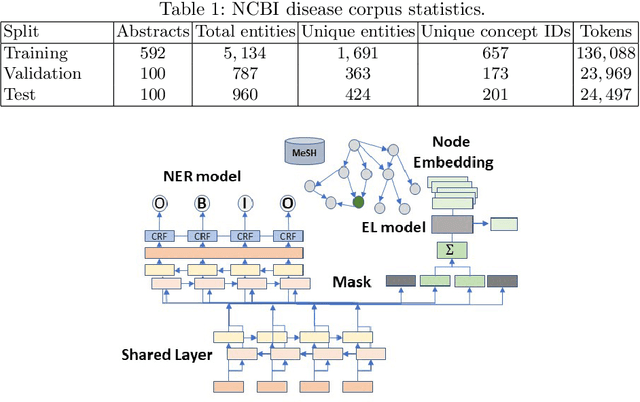
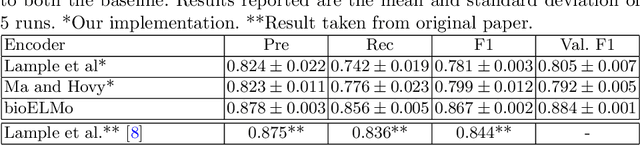
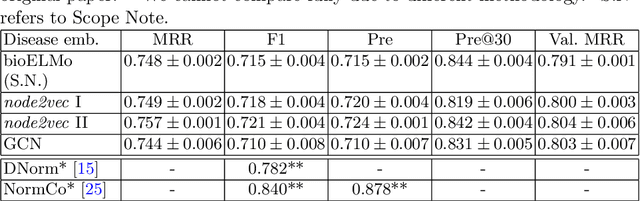
Abstract:The detection and normalization of diseases in biomedical texts are key biomedical natural language processing tasks. Disease names need not only be identified, but also normalized or linked to clinical taxonomies describing diseases such as MeSH. In this paper we describe deep learning methods that tackle both tasks. We train and test our methods on the known NCBI disease benchmark corpus. We propose to represent disease names by leveraging MeSH's graphical structure together with the lexical information available in the taxonomy using graph embeddings. We also show that combining neural named entity recognition models with our graph-based entity linking methods via multitask learning leads to improved disease recognition in the NCBI corpus.
Word Embeddings for Chemical Patent Natural Language Processing
Oct 24, 2020
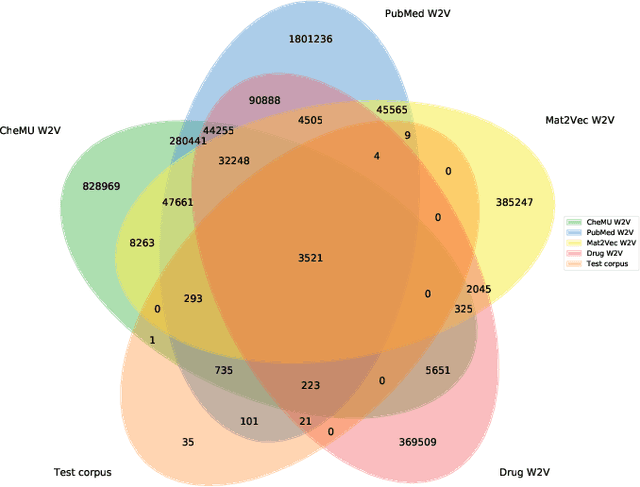
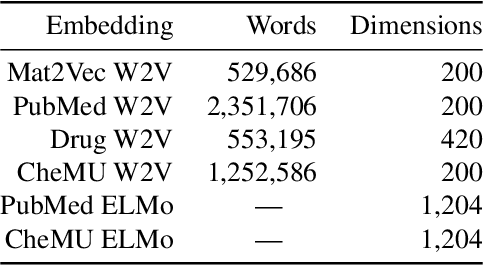
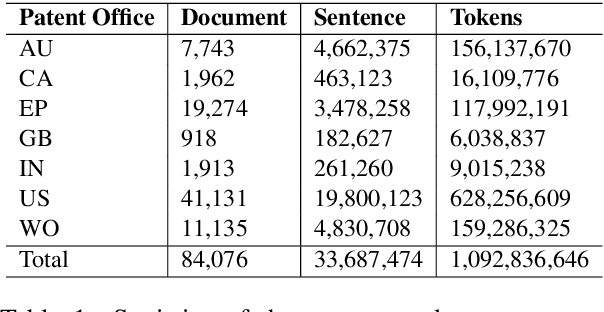
Abstract:We evaluate chemical patent word embeddings against known biomedical embeddings and show that they outperform the latter extrinsically and intrinsically. We also show that using contextualized embeddings can induce predictive models of reasonable performance for this domain over a relatively small gold standard.
Improving Chemical Named Entity Recognition in Patents with Contextualized Word Embeddings
Jul 05, 2019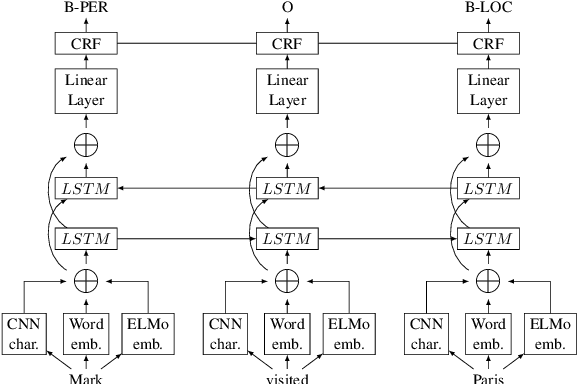
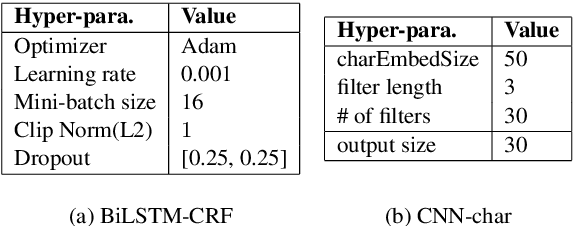
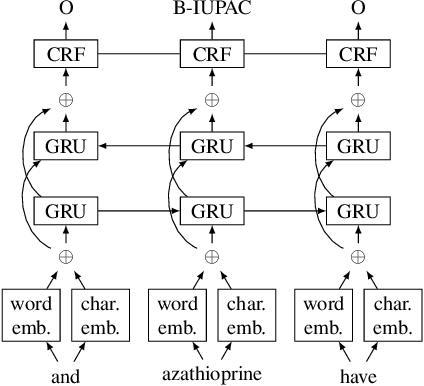
Abstract:Chemical patents are an important resource for chemical information. However, few chemical Named Entity Recognition (NER) systems have been evaluated on patent documents, due in part to their structural and linguistic complexity. In this paper, we explore the NER performance of a BiLSTM-CRF model utilising pre-trained word embeddings, character-level word representations and contextualized ELMo word representations for chemical patents. We compare word embeddings pre-trained on biomedical and chemical patent corpora. The effect of tokenizers optimized for the chemical domain on NER performance in chemical patents is also explored. The results on two patent corpora show that contextualized word representations generated from ELMo substantially improve chemical NER performance w.r.t. the current state-of-the-art. We also show that domain-specific resources such as word embeddings trained on chemical patents and chemical-specific tokenizers have a positive impact on NER performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge