Benoit Rosa
SAF-IS: a Spatial Annotation Free Framework for Instance Segmentation of Surgical Tools
Sep 04, 2023



Abstract:Instance segmentation of surgical instruments is a long-standing research problem, crucial for the development of many applications for computer-assisted surgery. This problem is commonly tackled via fully-supervised training of deep learning models, requiring expensive pixel-level annotations to train. In this work, we develop a framework for instance segmentation not relying on spatial annotations for training. Instead, our solution only requires binary tool masks, obtainable using recent unsupervised approaches, and binary tool presence labels, freely obtainable in robot-assisted surgery. Based on the binary mask information, our solution learns to extract individual tool instances from single frames, and to encode each instance into a compact vector representation, capturing its semantic features. Such representations guide the automatic selection of a tiny number of instances (8 only in our experiments), displayed to a human operator for tool-type labelling. The gathered information is finally used to match each training instance with a binary tool presence label, providing an effective supervision signal to train a tool instance classifier. We validate our framework on the EndoVis 2017 and 2018 segmentation datasets. We provide results using binary masks obtained either by manual annotation or as predictions of an unsupervised binary segmentation model. The latter solution yields an instance segmentation approach completely free from spatial annotations, outperforming several state-of-the-art fully-supervised segmentation approaches.
Semi-supervised GAN for Bladder Tissue Classification in Multi-Domain Endoscopic Images
Dec 21, 2022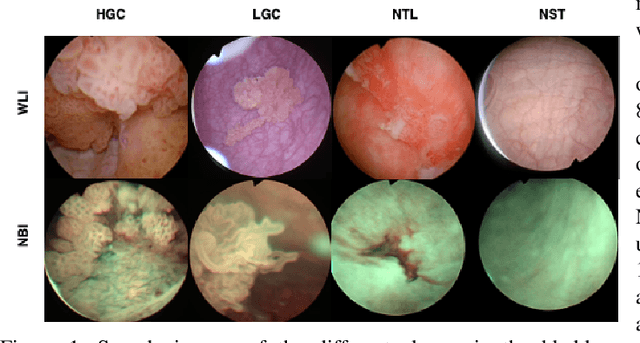
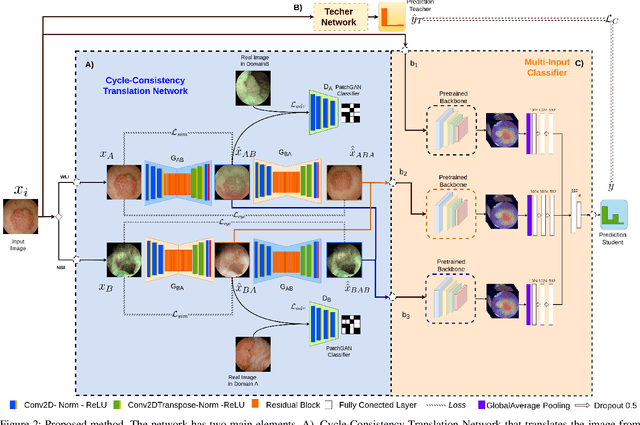
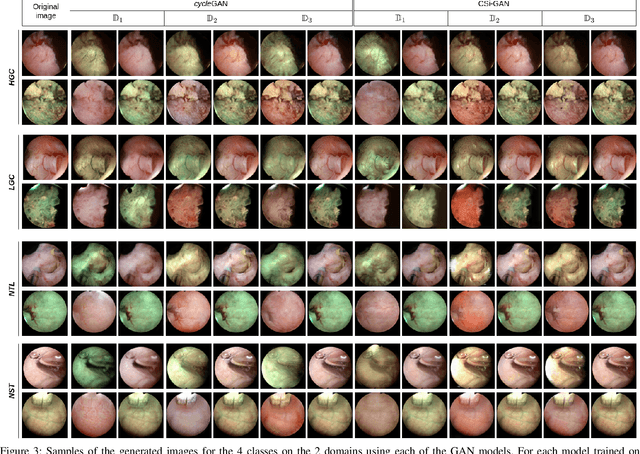
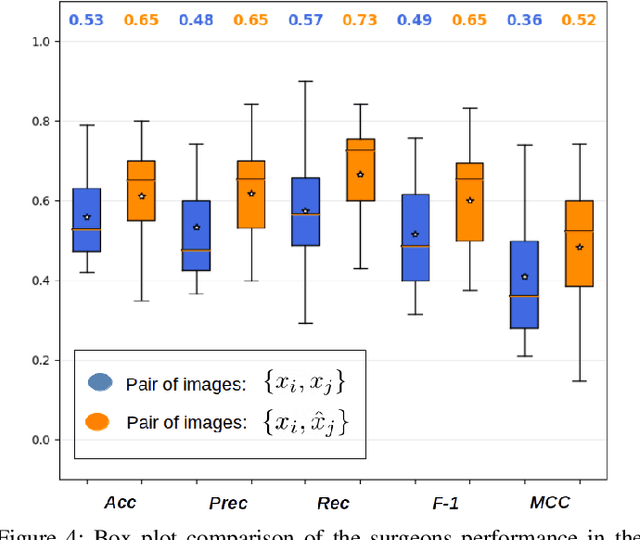
Abstract:Objective: Accurate visual classification of bladder tissue during Trans-Urethral Resection of Bladder Tumor (TURBT) procedures is essential to improve early cancer diagnosis and treatment. During TURBT interventions, White Light Imaging (WLI) and Narrow Band Imaging (NBI) techniques are used for lesion detection. Each imaging technique provides diverse visual information that allows clinicians to identify and classify cancerous lesions. Computer vision methods that use both imaging techniques could improve endoscopic diagnosis. We address the challenge of tissue classification when annotations are available only in one domain, in our case WLI, and the endoscopic images correspond to an unpaired dataset, i.e. there is no exact equivalent for every image in both NBI and WLI domains. Method: We propose a semi-surprised Generative Adversarial Network (GAN)-based method composed of three main components: a teacher network trained on the labeled WLI data; a cycle-consistency GAN to perform unpaired image-to-image translation, and a multi-input student network. To ensure the quality of the synthetic images generated by the proposed GAN we perform a detailed quantitative, and qualitative analysis with the help of specialists. Conclusion: The overall average classification accuracy, precision, and recall obtained with the proposed method for tissue classification are 0.90, 0.88, and 0.89 respectively, while the same metrics obtained in the unlabeled domain (NBI) are 0.92, 0.64, and 0.94 respectively. The quality of the generated images is reliable enough to deceive specialists. Significance: This study shows the potential of using semi-supervised GAN-based classification to improve bladder tissue classification when annotations are limited in multi-domain data.
Autonomous Intraluminal Navigation of a Soft Robot using Deep-Learning-based Visual Servoing
Jul 01, 2022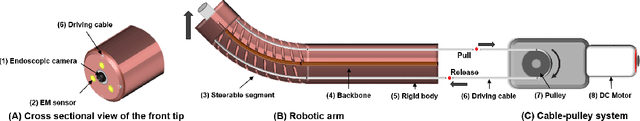
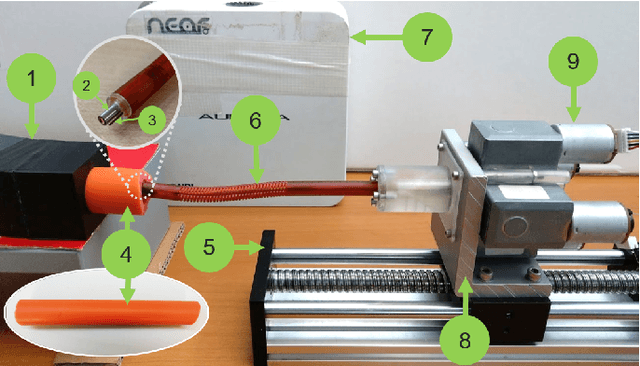
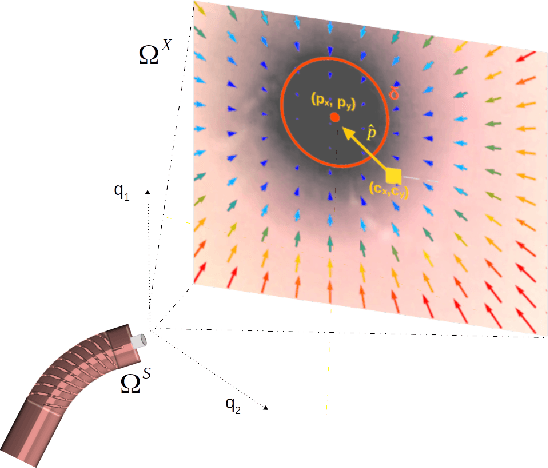
Abstract:Navigation inside luminal organs is an arduous task that requires non-intuitive coordination between the movement of the operator's hand and the information obtained from the endoscopic video. The development of tools to automate certain tasks could alleviate the physical and mental load of doctors during interventions, allowing them to focus on diagnosis and decision-making tasks. In this paper, we present a synergic solution for intraluminal navigation consisting of a 3D printed endoscopic soft robot that can move safely inside luminal structures. Visual servoing, based on Convolutional Neural Networks (CNNs) is used to achieve the autonomous navigation task. The CNN is trained with phantoms and in-vivo data to segment the lumen, and a model-less approach is presented to control the movement in constrained environments. The proposed robot is validated in anatomical phantoms in different path configurations. We analyze the movement of the robot using different metrics such as task completion time, smoothness, error in the steady-state, and mean and maximum error. We show that our method is suitable to navigate safely in hollow environments and conditions which are different than the ones the network was originally trained on.
FUN-SIS: a Fully UNsupervised approach for Surgical Instrument Segmentation
Feb 16, 2022



Abstract:Automatic surgical instrument segmentation of endoscopic images is a crucial building block of many computer-assistance applications for minimally invasive surgery. So far, state-of-the-art approaches completely rely on the availability of a ground-truth supervision signal, obtained via manual annotation, thus expensive to collect at large scale. In this paper, we present FUN-SIS, a Fully-UNsupervised approach for binary Surgical Instrument Segmentation. FUN-SIS trains a per-frame segmentation model on completely unlabelled endoscopic videos, by solely relying on implicit motion information and instrument shape-priors. We define shape-priors as realistic segmentation masks of the instruments, not necessarily coming from the same dataset/domain as the videos. The shape-priors can be collected in various and convenient ways, such as recycling existing annotations from other datasets. We leverage them as part of a novel generative-adversarial approach, allowing to perform unsupervised instrument segmentation of optical-flow images during training. We then use the obtained instrument masks as pseudo-labels in order to train a per-frame segmentation model; to this aim, we develop a learning-from-noisy-labels architecture, designed to extract a clean supervision signal from these pseudo-labels, leveraging their peculiar noise properties. We validate the proposed contributions on three surgical datasets, including the MICCAI 2017 EndoVis Robotic Instrument Segmentation Challenge dataset. The obtained fully-unsupervised results for surgical instrument segmentation are almost on par with the ones of fully-supervised state-of-the-art approaches. This suggests the tremendous potential of the proposed method to leverage the great amount of unlabelled data produced in the context of minimally invasive surgery.
Data Stream Stabilization for Optical Coherence Tomography Volumetric Scanning
Dec 02, 2021



Abstract:Optical Coherence Tomography (OCT) is an emerging medical imaging modality for luminal organ diagnosis. The non-constant rotation speed of optical components in the OCT catheter tip causes rotational distortion in OCT volumetric scanning. By improving the scanning process, this instability can be partially reduced. To further correct the rotational distortion in the OCT image, a volumetric data stabilization algorithm is proposed. The algorithm first estimates the Non-Uniform Rotational Distortion (NURD) for each B-scan by using a Convolutional Neural Network (CNN). A correlation map between two successive B-scans is computed and provided as input to the CNN. To solve the problem of accumulative error in iterative frame stream processing, we deploy an overall rotation estimation between reference orientation and actual OCT image orientation. We train the network with synthetic OCT videos by intentionally adding rotational distortion into real OCT images. As part of this article we discuss the proposed method in two different scanning modes: the first is a conventional pullback mode where the optical components move along the protection sheath, and the second is a self-designed scanning mode where the catheter is globally translated by using an external actuator. The efficiency and robustness of the proposed method are evaluated with synthetic scans as well as real scans under two scanning modes.
* 11pages, 5 figures
A transfer-learning approach for lesion detection in endoscopic images from the urinary tract
Apr 08, 2021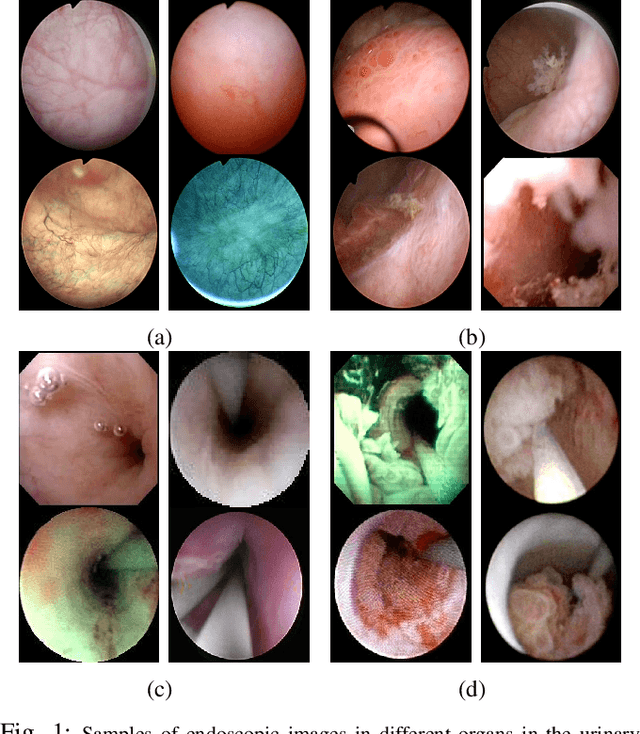
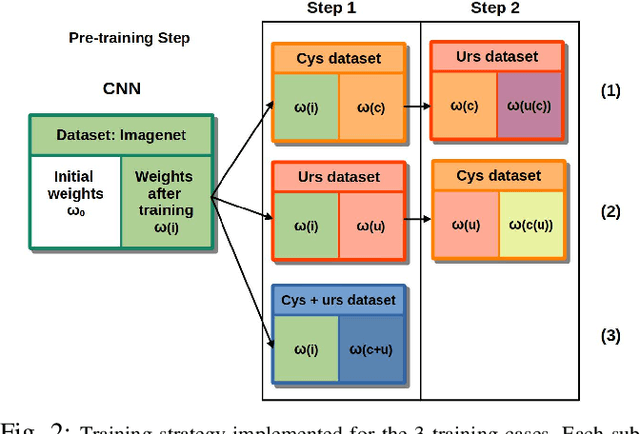
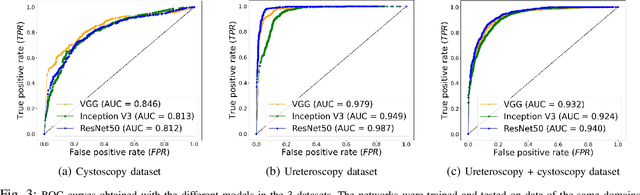
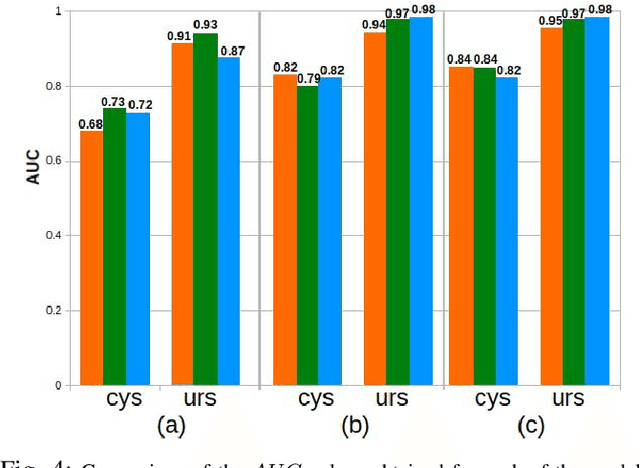
Abstract:Ureteroscopy and cystoscopy are the gold standard methods to identify and treat tumors along the urinary tract. It has been reported that during a normal procedure a rate of 10-20 % of the lesions could be missed. In this work we study the implementation of 3 different Convolutional Neural Networks (CNNs), using a 2-steps training strategy, to classify images from the urinary tract with and without lesions. A total of 6,101 images from ureteroscopy and cystoscopy procedures were collected. The CNNs were trained and tested using transfer learning in a two-steps fashion on 3 datasets. The datasets used were: 1) only ureteroscopy images, 2) only cystoscopy images and 3) the combination of both of them. For cystoscopy data, VGG performed better obtaining an Area Under the ROC Curve (AUC) value of 0.846. In the cases of ureteroscopy and the combination of both datasets, ResNet50 achieved the best results with AUC values of 0.987 and 0.940. The use of a training dataset that comprehends both domains results in general better performances, but performing a second stage of transfer learning achieves comparable ones. There is no single model which performs better in all scenarios, but ResNet50 is the network that achieves the best performances in most of them. The obtained results open the opportunity for further investigation with a view for improving lesion detection in endoscopic images of the urinary system.
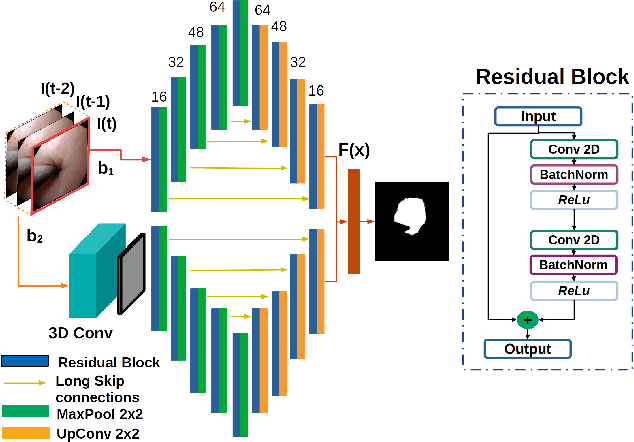
Using spatial-temporal ensembles of convolutional neural networks for lumen segmentation in ureteroscopy
Apr 05, 2021



Abstract:Purpose: Ureteroscopy is an efficient endoscopic minimally invasive technique for the diagnosis and treatment of upper tract urothelial carcinoma (UTUC). During ureteroscopy, the automatic segmentation of the hollow lumen is of primary importance, since it indicates the path that the endoscope should follow. In order to obtain an accurate segmentation of the hollow lumen, this paper presents an automatic method based on Convolutional Neural Networks (CNNs). Methods: The proposed method is based on an ensemble of 4 parallel CNNs to simultaneously process single and multi-frame information. Of these, two architectures are taken as core-models, namely U-Net based in residual blocks($m_1$) and Mask-RCNN($m_2$), which are fed with single still-frames $I(t)$. The other two models ($M_1$, $M_2$) are modifications of the former ones consisting on the addition of a stage which makes use of 3D Convolutions to process temporal information. $M_1$, $M_2$ are fed with triplets of frames ($I(t-1)$, $I(t)$, $I(t+1)$) to produce the segmentation for $I(t)$. Results: The proposed method was evaluated using a custom dataset of 11 videos (2,673 frames) which were collected and manually annotated from 6 patients. We obtain a Dice similarity coefficient of 0.80, outperforming previous state-of-the-art methods. Conclusion: The obtained results show that spatial-temporal information can be effectively exploited by the ensemble model to improve hollow lumen segmentation in ureteroscopic images. The method is effective also in presence of poor visibility, occasional bleeding, or specular reflections.
A Kinematic Bottleneck Approach For Pose Regression of Flexible Surgical Instruments directly from Images
Feb 28, 2021



Abstract:3-D pose estimation of instruments is a crucial step towards automatic scene understanding in robotic minimally invasive surgery. Although robotic systems can potentially directly provide joint values, this information is not commonly exploited inside the operating room, due to its possible unreliability, limited access and the time-consuming calibration required, especially for continuum robots. For this reason, standard approaches for 3-D pose estimation involve the use of external tracking systems. Recently, image-based methods have emerged as promising, non-invasive alternatives. While many image-based approaches in the literature have shown accurate results, they generally require either a complex iterative optimization for each processed image, making them unsuitable for real-time applications, or a large number of manually-annotated images for efficient learning. In this paper we propose a self-supervised image-based method, exploiting, at training time only, the imprecise kinematic information provided by the robot. In order to avoid introducing time-consuming manual annotations, the problem is formulated as an auto-encoder, smartly bottlenecked by the presence of a physical model of the robotic instruments and surgical camera, forcing a separation between image background and kinematic content. Validation of the method was performed on semi-synthetic, phantom and in-vivo datasets, obtained using a flexible robotized endoscope, showing promising results for real-time image-based 3-D pose estimation of surgical instruments.
A Lumen Segmentation Method in Ureteroscopy Images based on a Deep Residual U-Net architecture
Jan 13, 2021



Abstract:Ureteroscopy is becoming the first surgical treatment option for the majority of urinary affections. This procedure is performed using an endoscope which provides the surgeon with the visual information necessary to navigate inside the urinary tract. Having in mind the development of surgical assistance systems, that could enhance the performance of surgeon, the task of lumen segmentation is a fundamental part since this is the visual reference which marks the path that the endoscope should follow. This is something that has not been analyzed in ureteroscopy data before. However, this task presents several challenges given the image quality and the conditions itself of ureteroscopy procedures. In this paper, we study the implementation of a Deep Neural Network which exploits the advantage of residual units in an architecture based on U-Net. For the training of these networks, we analyze the use of two different color spaces: gray-scale and RGB data images. We found that training on gray-scale images gives the best results obtaining mean values of Dice Score, Precision, and Recall of 0.73, 0.58, and 0.92 respectively. The results obtained shows that the use of residual U-Net could be a suitable model for further development for a computer-aided system for navigation and guidance through the urinary system.
Self-Supervised Surgical Tool Segmentation using Kinematic Information
Feb 13, 2019



Abstract:Surgical tool segmentation in endoscopic images is the first step towards pose estimation and (sub-)task automation in challenging minimally invasive surgical operations. While many approaches in the literature have shown great results using modern machine learning methods such as convolutional neural networks, the main bottleneck lies in the acquisition of a large number of manually-annotated images for efficient learning. This is especially true in surgical context, where patient-to-patient differences impede the overall generalizability. In order to cope with this lack of annotated data, we propose a self-supervised approach in a robot-assisted context. To our knowledge, the proposed approach is the first to make use of the kinematic model of the robot in order to generate training labels. The core contribution of the paper is to propose an optimization method to obtain good labels for training despite an unknown hand-eye calibration and an imprecise kinematic model. The labels can subsequently be used for fine-tuning a fully-convolutional neural network for pixel-wise classification. As a result, the tool can be segmented in the endoscopic images without needing a single manually-annotated image. Experimental results on phantom and in vivo datasets obtained using a flexible robotized endoscopy system are very promising.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge