Aurelien Bustin
Deep Learning Based Detection and Correction of Cardiac MR Motion Artefacts During Reconstruction for High-Quality Segmentation
Oct 21, 2019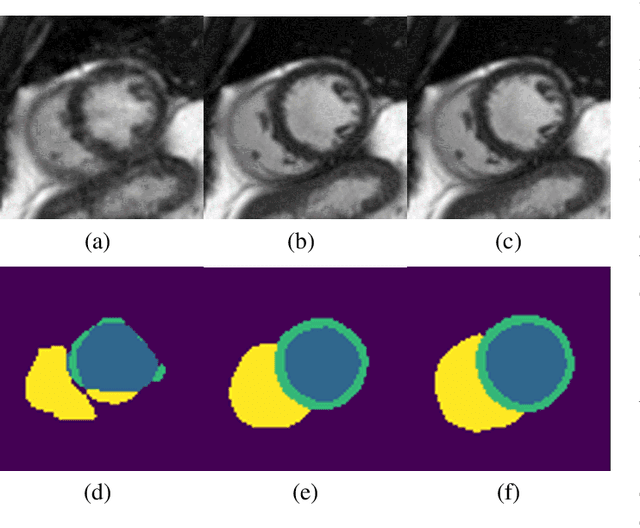
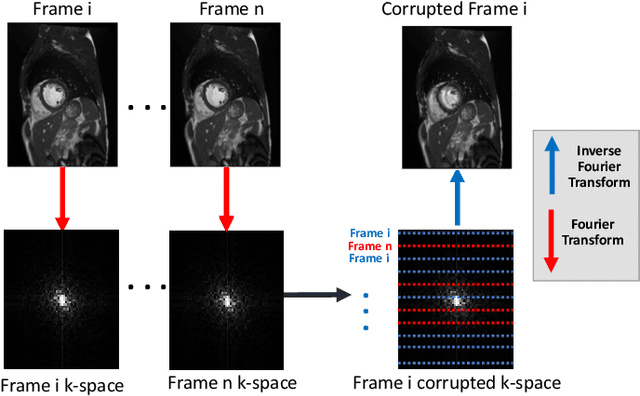
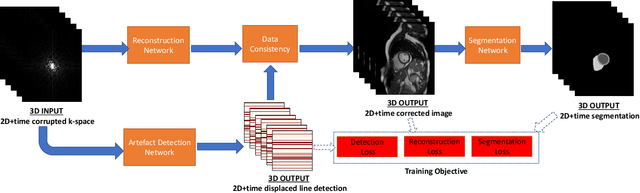
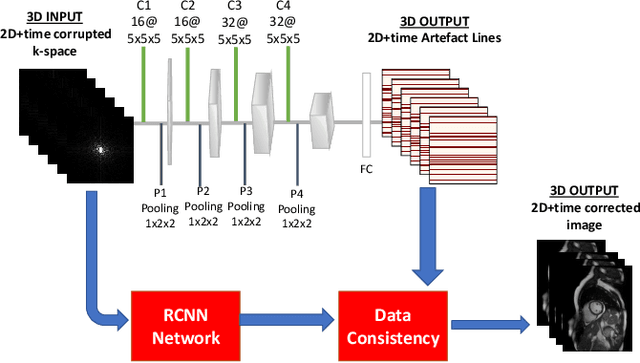
Abstract:Segmenting anatomical structures in medical images has been successfully addressed with deep learning methods for a range of applications. However, this success is heavily dependent on the quality of the image that is being segmented. A commonly neglected point in the medical image analysis community is the vast amount of clinical images that have severe image artefacts due to organ motion, movement of the patient and/or image acquisition related issues. In this paper, we discuss the implications of image motion artefacts on cardiac MR segmentation and compare a variety of approaches for jointly correcting for artefacts and segmenting the cardiac cavity. We propose to use a segmentation network coupled with this in an end-to-end framework. Our training optimises three different tasks: 1) image artefact detection, 2) artefact correction and 3) image segmentation. We train the reconstruction network to automatically correct for motion-related artefacts using synthetically corrupted cardiac MR k-space data and uncorrected reconstructed images. Using a test set of 500 2D+time cine MR acquisitions from the UK Biobank data set, we achieve demonstrably good image quality and high segmentation accuracy in the presence of synthetic motion artefacts. We quantitatively compare our method with a variety of techniques for jointly recovering image quality and performing image segmentation. We showcase better performance compared to state-of-the-art image correction techniques. Moreover, our method preserves the quality of uncorrupted images and therefore can be utilised as a global image reconstruction algorithm.
Detection and Correction of Cardiac MR Motion Artefacts during Reconstruction from K-space
Jun 12, 2019



Abstract:In fully sampled cardiac MR (CMR) acquisitions, motion can lead to corruption of k-space lines, which can result in artefacts in the reconstructed images. In this paper, we propose a method to automatically detect and correct motion-related artefacts in CMR acquisitions during reconstruction from k-space data. Our correction method is inspired by work on undersampled CMR reconstruction, and uses deep learning to optimize a data-consistency term for under-sampled k-space reconstruction. Our main methodological contribution is the addition of a detection network to classify motion-corrupted k-space lines to convert the problem of artefact correction to a problem of reconstruction using the data consistency term. We train our network to automatically correct for motion-related artefacts using synthetically corrupted cine CMR k-space data as well as uncorrupted CMR images. Using a test set of 50 2D+time cine CMR datasets from the UK Biobank, we achieve good image quality in the presence of synthetic motion artefacts. We quantitatively compare our method with a variety of techniques for recovering good image quality and showcase better performance compared to state of the art denoising techniques with a PSNR of 37.1. Moreover, we show that our method preserves the quality of uncorrupted images and therefore can be also utilized as a general image reconstruction algorithm.
Magnetic Resonance Fingerprinting using Recurrent Neural Networks
Dec 19, 2018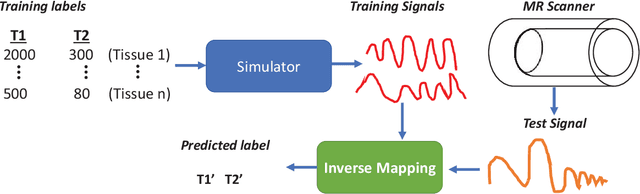
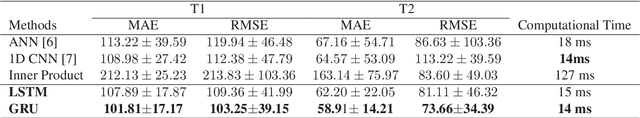
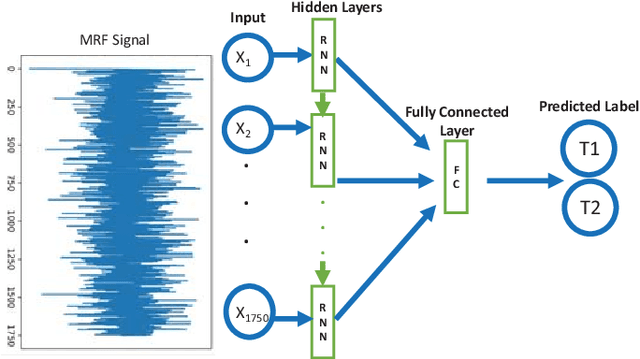
Abstract:Magnetic Resonance Fingerprinting (MRF) is a new approach to quantitative magnetic resonance imaging that allows simultaneous measurement of multiple tissue properties in a single, time-efficient acquisition. Standard MRF reconstructs parametric maps using dictionary matching and lacks scalability due to computational inefficiency. We propose to perform MRF map reconstruction using a recurrent neural network, which exploits the time-dependent information of the MRF signal evolution. We evaluate our method on multiparametric synthetic signals and compare it to existing MRF map reconstruction approaches, including those based on neural networks. Our method achieves state-of-the-art estimates of T1 and T2 values. In addition, the reconstruction time is significantly reduced compared to dictionary-matching based approaches.
Automatic CNN-based detection of cardiac MR motion artefacts using k-space data augmentation and curriculum learning
Oct 29, 2018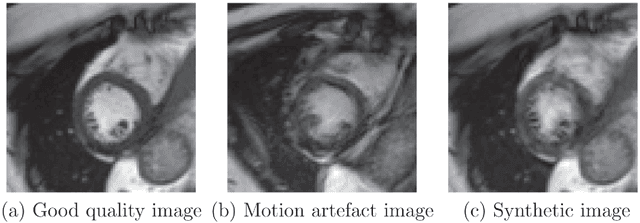
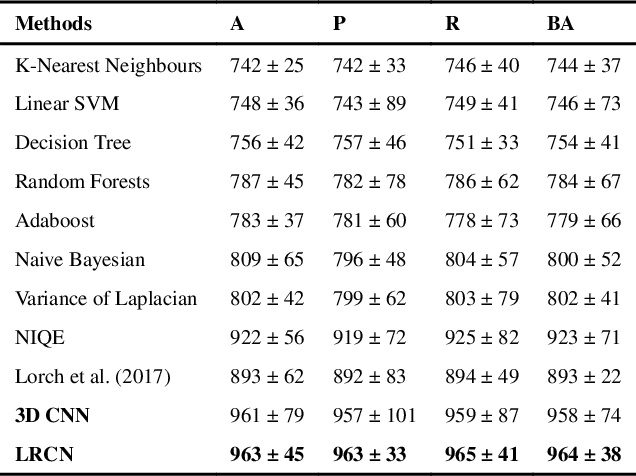

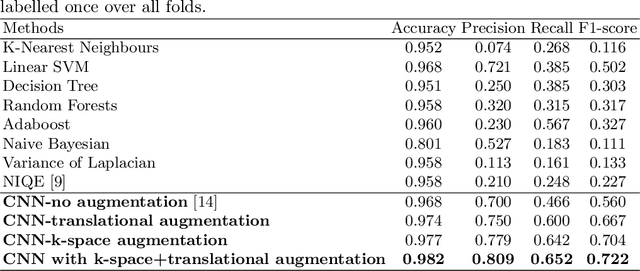
Abstract:Good quality of medical images is a prerequisite for the success of subsequent image analysis pipelines. Quality assessment of medical images is therefore an essential activity and for large population studies such as the UK Biobank (UKBB), manual identification of artefacts such as those caused by unanticipated motion is tedious and time-consuming. Therefore, there is an urgent need for automatic image quality assessment techniques. In this paper, we propose a method to automatically detect the presence of motion-related artefacts in cardiac magnetic resonance (CMR) cine images. We compare two deep learning architectures to classify poor quality CMR images: 1) 3D spatio-temporal Convolutional Neural Networks (3D-CNN), 2) Long-term Recurrent Convolutional Network (LRCN). Though in real clinical setup motion artefacts are common, high-quality imaging of UKBB, which comprises cross-sectional population data of volunteers who do not necessarily have health problems creates a highly imbalanced classification problem. Due to the high number of good quality images compared to the relatively low number of images with motion artefacts, we propose a novel data augmentation scheme based on synthetic artefact creation in k-space. We also investigate a learning approach using a predetermined curriculum based on synthetic artefact severity. We evaluate our pipeline on a subset of the UK Biobank data set consisting of 3510 CMR images. The LRCN architecture outperformed the 3D-CNN architecture and was able to detect 2D+time short axis images with motion artefacts in less than 1ms with high recall. We compare our approach to a range of state-of-the-art quality assessment methods. The novel data augmentation and curriculum learning approaches both improved classification performance achieving overall area under the ROC curve of 0.89.
Deep Learning using K-space Based Data Augmentation for Automated Cardiac MR Motion Artefact Detection
Aug 31, 2018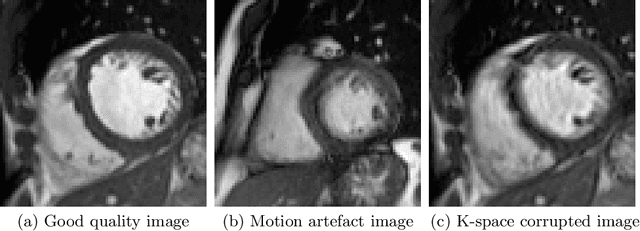
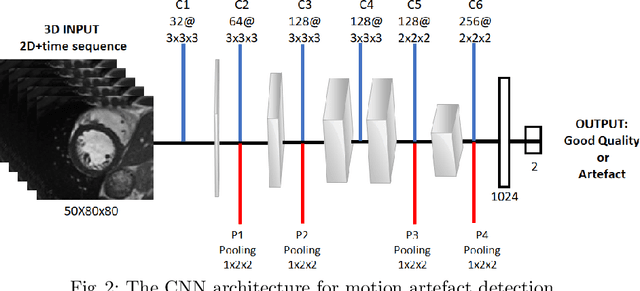
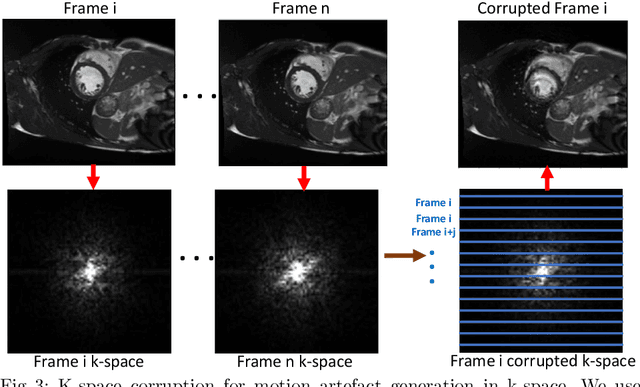
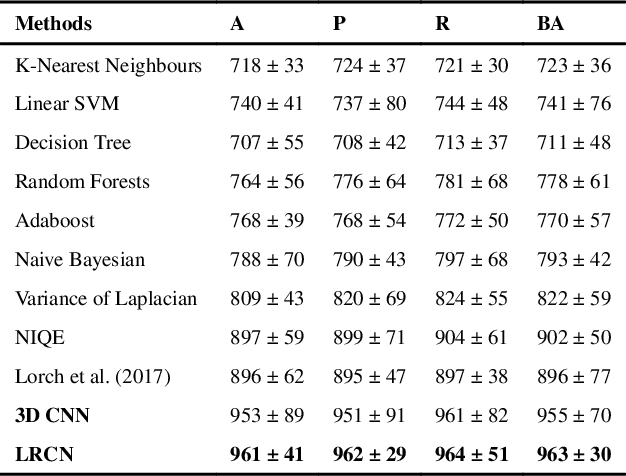
Abstract:Quality assessment of medical images is essential for complete automation of image processing pipelines. For large population studies such as the UK Biobank, artefacts such as those caused by heart motion are problematic and manual identification is tedious and time-consuming. Therefore, there is an urgent need for automatic image quality assessment techniques. In this paper, we propose a method to automatically detect the presence of motion-related artefacts in cardiac magnetic resonance (CMR) images. As this is a highly imbalanced classification problem (due to the high number of good quality images compared to the low number of images with motion artefacts), we propose a novel k-space based training data augmentation approach in order to address this problem. Our method is based on 3D spatio-temporal Convolutional Neural Networks, and is able to detect 2D+time short axis images with motion artefacts in less than 1ms. We test our algorithm on a subset of the UK Biobank dataset consisting of 3465 CMR images and achieve not only high accuracy in detection of motion artefacts, but also high precision and recall. We compare our approach to a range of state-of-the-art quality assessment methods.
Motion Estimated-Compensated Reconstruction with Preserved-Features in Free-Breathing Cardiac MRI
Nov 15, 2016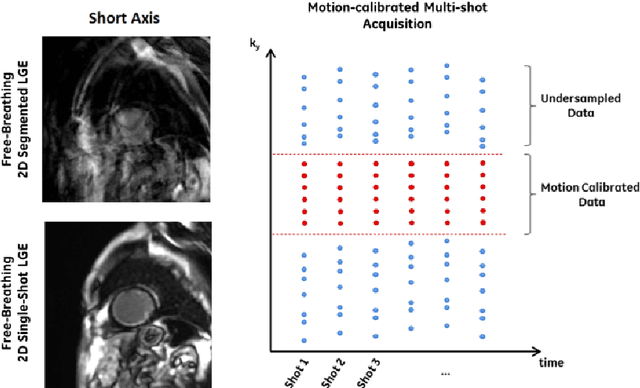
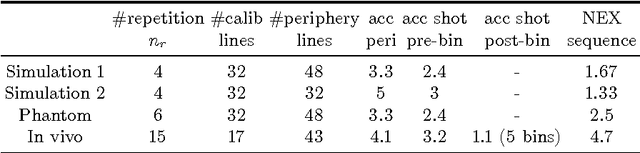
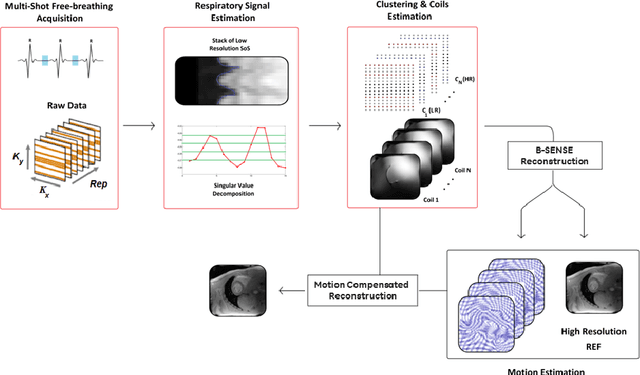
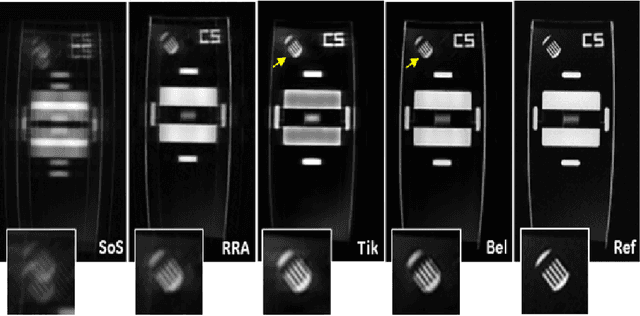
Abstract:To develop an efficient motion-compensated reconstruction technique for free-breathing cardiac magnetic resonance imaging (MRI) that allows high-quality images to be reconstructed from multiple undersampled single-shot acquisitions. The proposed method is a joint image reconstruction and motion correction method consisting of several steps, including a non-rigid motion extraction and a motion-compensated reconstruction. The reconstruction includes a denoising with the Beltrami regularization, which offers an ideal compromise between feature preservation and staircasing reduction. Results were assessed in simulation, phantom and volunteer experiments. The proposed joint image reconstruction and motion correction method exhibits visible quality improvement over previous methods while reconstructing sharper edges. Moreover, when the acceleration factor increases, standard methods show blurry results while the proposed method preserves image quality. The method was applied to free-breathing single-shot cardiac MRI, successfully achieving high image quality and higher spatial resolution than conventional segmented methods, with the potential to offer high-quality delayed enhancement scans in challenging patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge