Assaf Hoogi
Unsupervised Iterative U-Net with an Internal Guidance Layer for Vertebrae Contrast Enhancement in Chest X-Ray Images
Jun 06, 2023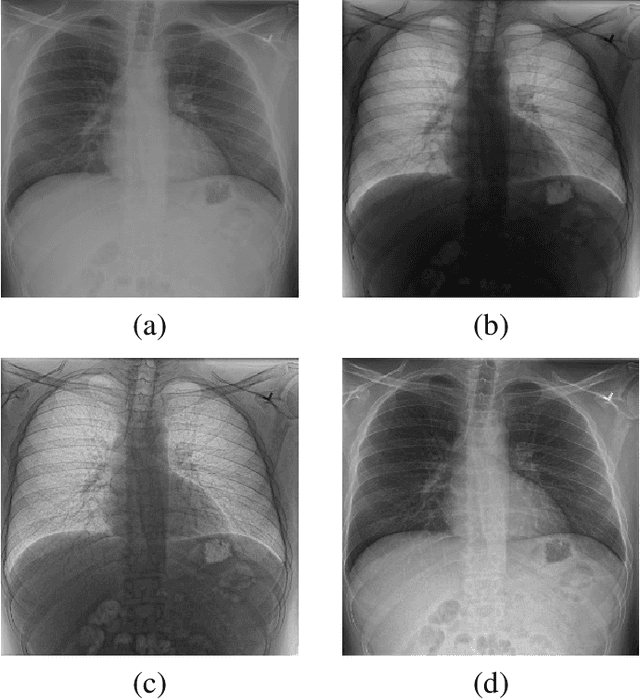
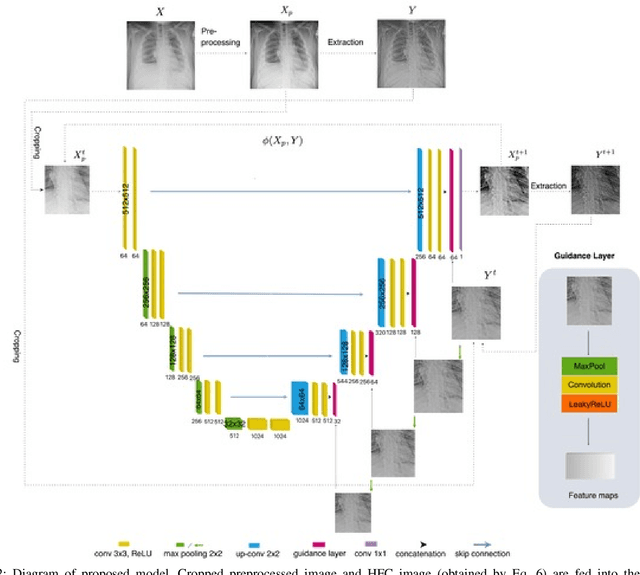
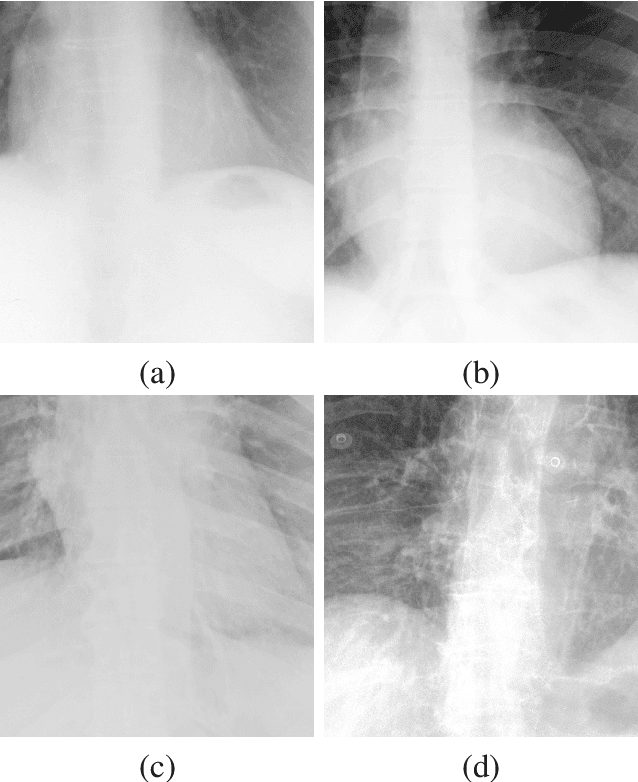
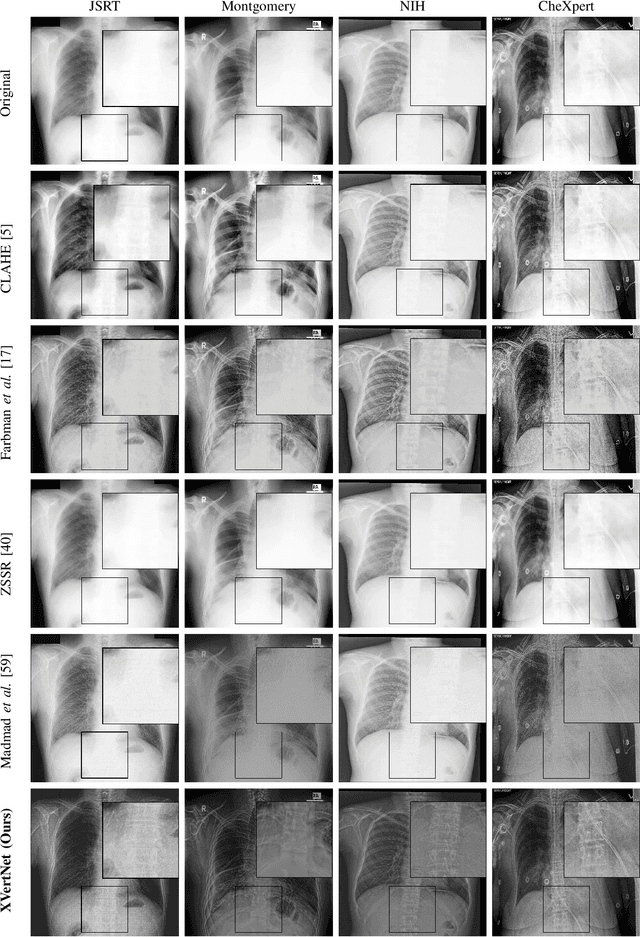
Abstract:X-ray imaging is a fundamental clinical tool for screening and diagnosing various diseases. However, the spatial resolution of radiographs is often limited, making it challenging to diagnose small image details and leading to difficulties in identifying vertebrae anomalies at an early stage in chest radiographs. To address this limitation, we propose a novel and robust approach to significantly improve the quality of X-ray images by iteratively training a deep neural network. Our framework includes an embedded internal guidance layer that enhances the fine structures of spinal vertebrae in chest X-ray images through fully unsupervised training, utilizing an iterative procedure that employs the same network architecture in each enhancement phase. Additionally, we have designed an optimized loss function that accurately identifies object boundaries and enhances spinal features, thereby further enhancing the quality of the images. Experimental results demonstrate that our proposed method surpasses existing detail enhancement methods in terms of BRISQUE scores, and is comparable in terms of LPC-SI. Furthermore, our approach exhibits superior performance in restoring hidden fine structures, as evidenced by our qualitative results. This innovative approach has the potential to significantly enhance the diagnostic accuracy and early detection of diseases, making it a promising advancement in X-ray imaging technology.
Leveraging the Triple Exponential Moving Average for Fast-Adaptive Moment Estimation
Jun 02, 2023Abstract:Network optimization is a crucial step in the field of deep learning, as it directly affects the performance of models in various domains such as computer vision. Despite the numerous optimizers that have been developed over the years, the current methods are still limited in their ability to accurately and quickly identify gradient trends, which can lead to sub-optimal network performance. In this paper, we propose a novel deep optimizer called Fast-Adaptive Moment Estimation (FAME), which for the first time estimates gradient moments using a Triple Exponential Moving Average (TEMA). Incorporating TEMA into the optimization process provides richer and more accurate information on data changes and trends, as compared to the standard Exponential Moving Average used in essentially all current leading adaptive optimization methods. Our proposed FAME optimizer has been extensively validated through a wide range of benchmarks, including CIFAR-10, CIFAR-100, PASCAL-VOC, MS-COCO, and Cityscapes, using 14 different learning architectures, six optimizers, and various vision tasks, including detection, classification and semantic understanding. The results demonstrate that our FAME optimizer outperforms other leading optimizers in terms of both robustness and accuracy.
Deep Active Lesion Segmentation
Aug 21, 2019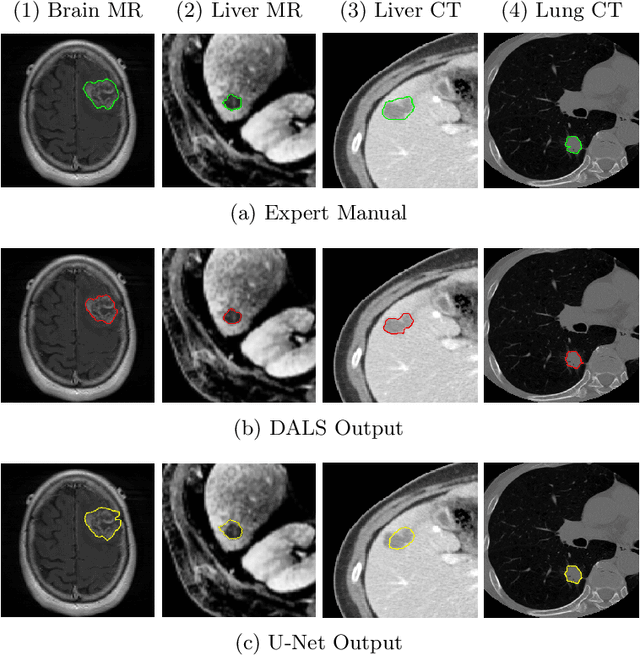
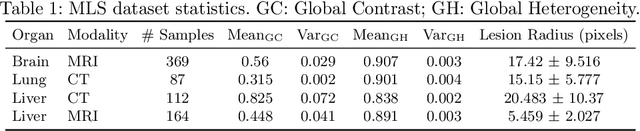
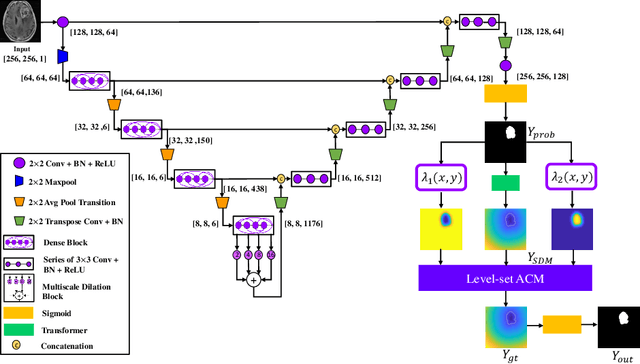
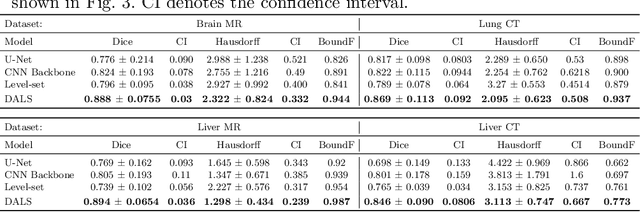
Abstract:Lesion segmentation is an important problem in computer-assisted diagnosis that remains challenging due to the prevalence of low contrast, irregular boundaries that are unamenable to shape priors. We introduce Deep Active Lesion Segmentation (DALS), a fully automated segmentation framework for that leverages the powerful nonlinear feature extraction abilities of fully Convolutional Neural Networks (CNNs) and the precise boundary delineation abilities of Active Contour Models (ACMs). Our DALS framework benefits from an improved level-set ACM formulation with a per-pixel-parameterized energy functional and a novel multiscale encoder-decoder CNN that learns an initialization probability map along with parameter maps for the ACM. We evaluate our lesion segmentation model on a new Multiorgan Lesion Segmentation (MLS) dataset that contains images of various organs, including brain, liver, and lung, across different imaging modalities---MR and CT. Our results demonstrate favorable performance compared to competing methods, especially for small training datasets.
* Accepted to Machine Learning in Medical Imaging (MLMI 2019)
From voxels to pixels and back: Self-supervision in natural-image reconstruction from fMRI
Jul 03, 2019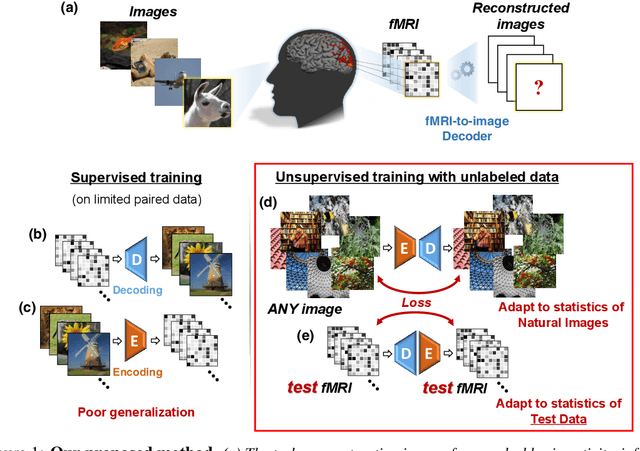
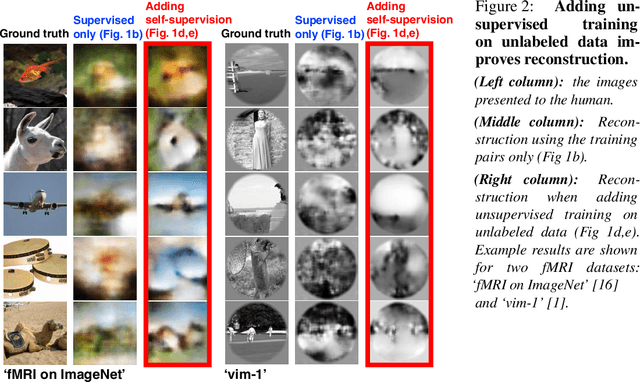
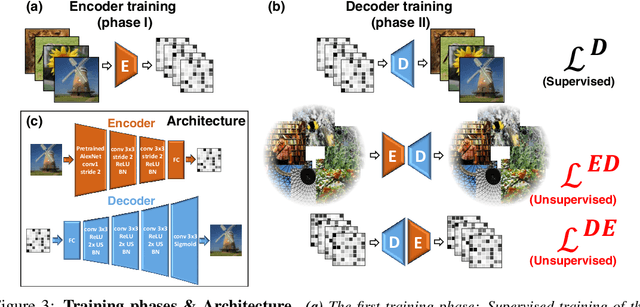
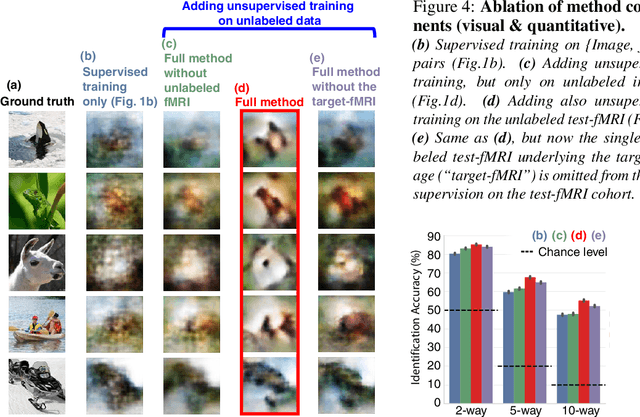
Abstract:Reconstructing observed images from fMRI brain recordings is challenging. Unfortunately, acquiring sufficient "labeled" pairs of {Image, fMRI} (i.e., images with their corresponding fMRI responses) to span the huge space of natural images is prohibitive for many reasons. We present a novel approach which, in addition to the scarce labeled data (training pairs), allows to train fMRI-to-image reconstruction networks also on "unlabeled" data (i.e., images without fMRI recording, and fMRI recording without images). The proposed model utilizes both an Encoder network (image-to-fMRI) and a Decoder network (fMRI-to-image). Concatenating these two networks back-to-back (Encoder-Decoder & Decoder-Encoder) allows augmenting the training with both types of unlabeled data. Importantly, it allows training on the unlabeled test-fMRI data. This self-supervision adapts the reconstruction network to the new input test-data, despite its deviation from the statistics of the scarce training data.
Self-Attention Capsule Networks for Image Classification
Apr 29, 2019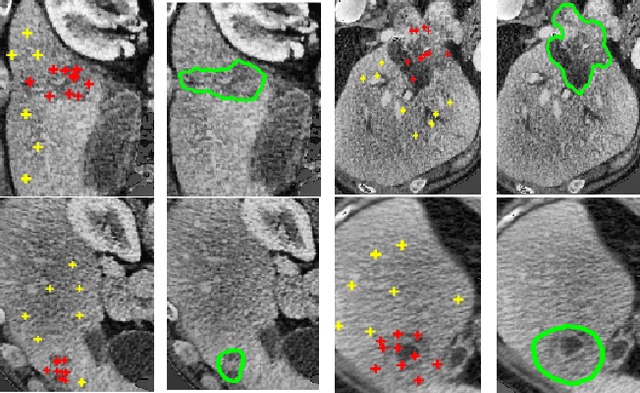
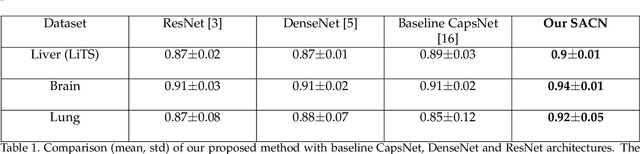
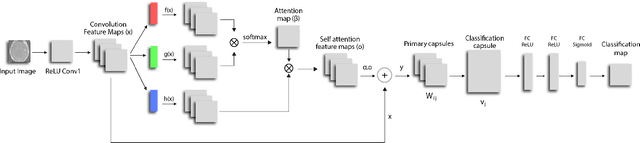
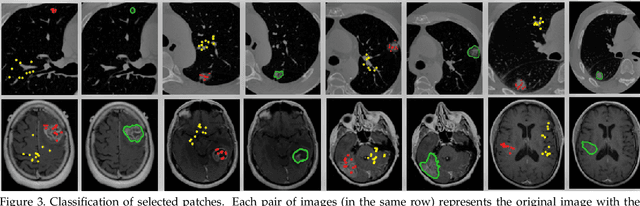
Abstract:We propose a novel architecture for image classification, called Self-Attention Capsule Networks (SACN). SACN is the first model that incorporates the Self-Attention mechanism as an integral layer within the Capsule Network (CapsNet). While the Self-Attention mechanism selects the more dominant image regions to focus on, the CapsNet analyzes the relevant features and their spatial correlations inside these regions only. The features are extracted in the convolutional layer. Then, the Self-Attention layer learns to suppress irrelevant regions based on features analysis, and highlights salient features useful for a specific task. The attention map is then fed into the CapsNet primary layer that is followed by a classification layer. The SACN proposed model was designed to use a relatively shallow CapsNet architecture to reduce computational load, and compensates for the absence of a deeper network by using the Self-Attention module to significantly improve the results. The proposed Self-Attention CapsNet architecture was extensively evaluated on five different datasets, mainly on three different medical sets, in addition to the natural MNIST and SVHN. The model was able to classify images and their patches with diverse and complex backgrounds better than the baseline CapsNet. As a result, the proposed Self-Attention CapsNet significantly improved classification performance within and across different datasets and outperformed the baseline CapsNet not only in classification accuracy but also in robustness.
TVAE: Triplet-Based Variational Autoencoder using Metric Learning
Apr 03, 2018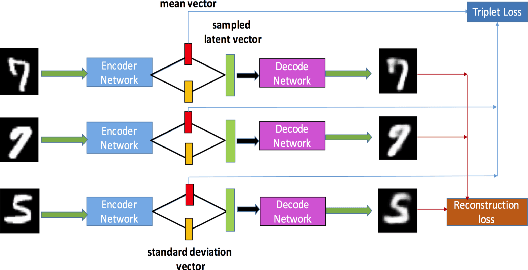
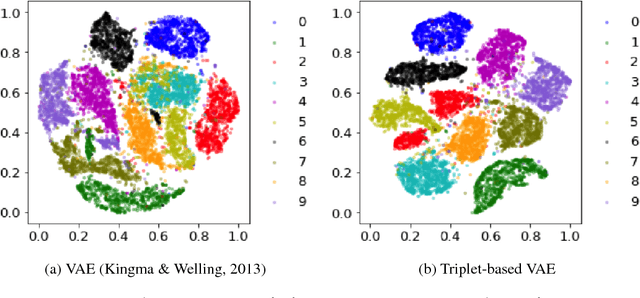

Abstract:Deep metric learning has been demonstrated to be highly effective in learning semantic representation and encoding information that can be used to measure data similarity, by relying on the embedding learned from metric learning. At the same time, variational autoencoder (VAE) has widely been used to approximate inference and proved to have a good performance for directed probabilistic models. However, for traditional VAE, the data label or feature information are intractable. Similarly, traditional representation learning approaches fail to represent many salient aspects of the data. In this project, we propose a novel integrated framework to learn latent embedding in VAE by incorporating deep metric learning. The features are learned by optimizing a triplet loss on the mean vectors of VAE in conjunction with standard evidence lower bound (ELBO) of VAE. This approach, which we call Triplet based Variational Autoencoder (TVAE), allows us to capture more fine-grained information in the latent embedding. Our model is tested on MNIST data set and achieves a high triplet accuracy of 95.60% while the traditional VAE (Kingma & Welling, 2013) achieves triplet accuracy of 75.08%.
A Fully-Automated Pipeline for Detection and Segmentation of Liver Lesions and Pathological Lymph Nodes
Mar 19, 2017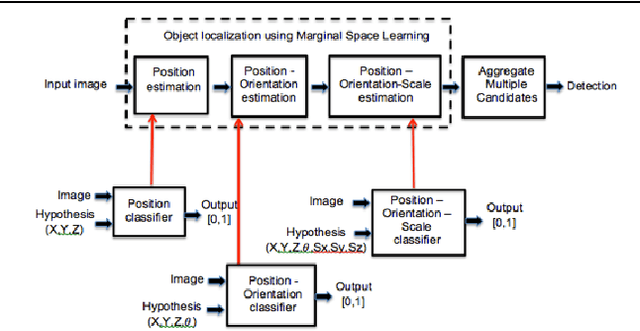

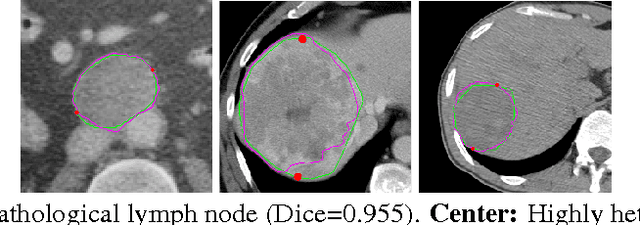
Abstract:We propose a fully-automated method for accurate and robust detection and segmentation of potentially cancerous lesions found in the liver and in lymph nodes. The process is performed in three steps, including organ detection, lesion detection and lesion segmentation. Our method applies machine learning techniques such as marginal space learning and convolutional neural networks, as well as active contour models. The method proves to be robust in its handling of extremely high lesion diversity. We tested our method on volumetric computed tomography (CT) images, including 42 volumes containing liver lesions and 86 volumes containing 595 pathological lymph nodes. Preliminary results under 10-fold cross validation show that for both the liver lesions and the lymph nodes, a total detection sensitivity of 0.53 and average Dice score of $0.71 \pm 0.15$ for segmentation were obtained.
Adaptive Local Window for Level Set Segmentation of CT and MRI Liver Lesions
Jun 12, 2016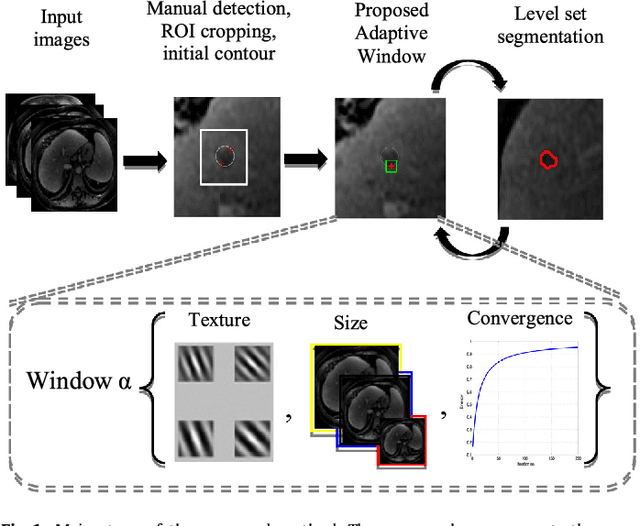
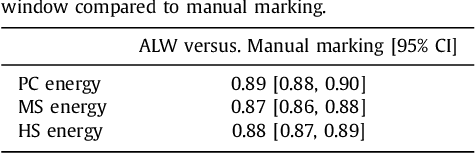
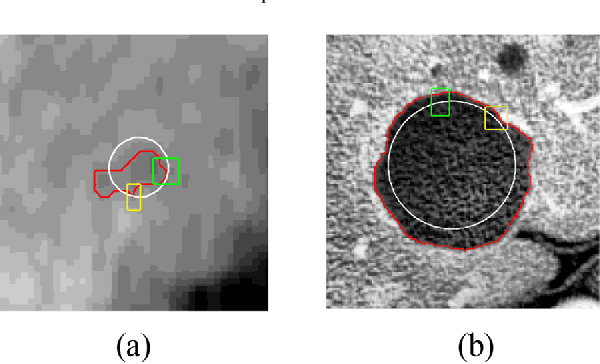
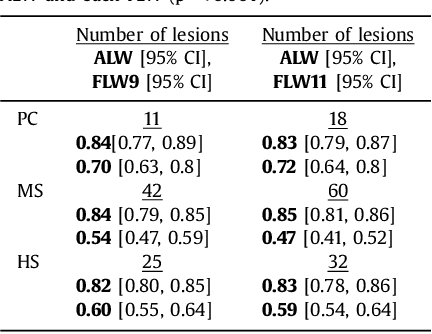
Abstract:We propose a novel method, the adaptive local window, for improving level set segmentation technique. The window is estimated separately for each contour point, over iterations of the segmentation process, and for each individual object. Our method considers the object scale, the spatial texture, and changes of the energy functional over iterations. Global and local statistics are considered by calculating several gray level co-occurrence matrices. We demonstrate the capabilities of the method in the domain of medical imaging for segmenting 233 images with liver lesions. To illustrate the strength of our method, those images were obtained by either Computed Tomography or Magnetic Resonance Imaging. Moreover, we analyzed images using three different energy models. We compare our method to a global level set segmentation and to local framework that uses predefined fixed-size square windows. The results indicate that our proposed method outperforms the other methods in terms of agreement with the manual marking and dependence on contour initialization or the energy model used. In case of complex lesions, such as low contrast lesions, heterogeneous lesions, or lesions with a noisy background, our method shows significantly better segmentation with an improvement of 0.25+- 0.13 in Dice similarity coefficient, compared with state of the art fixed-size local windows (Wilcoxon, p < 0.001).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge