Amir Ali Rahsepar
Shifts in Doctors' Eye Movements Between Real and AI-Generated Medical Images
Apr 21, 2025Abstract:Eye-tracking analysis plays a vital role in medical imaging, providing key insights into how radiologists visually interpret and diagnose clinical cases. In this work, we first analyze radiologists' attention and agreement by measuring the distribution of various eye-movement patterns, including saccades direction, amplitude, and their joint distribution. These metrics help uncover patterns in attention allocation and diagnostic strategies. Furthermore, we investigate whether and how doctors' gaze behavior shifts when viewing authentic (Real) versus deep-learning-generated (Fake) images. To achieve this, we examine fixation bias maps, focusing on first, last, short, and longest fixations independently, along with detailed saccades patterns, to quantify differences in gaze distribution and visual saliency between authentic and synthetic images.
Eyes Tell the Truth: GazeVal Highlights Shortcomings of Generative AI in Medical Imaging
Mar 26, 2025



Abstract:The demand for high-quality synthetic data for model training and augmentation has never been greater in medical imaging. However, current evaluations predominantly rely on computational metrics that fail to align with human expert recognition. This leads to synthetic images that may appear realistic numerically but lack clinical authenticity, posing significant challenges in ensuring the reliability and effectiveness of AI-driven medical tools. To address this gap, we introduce GazeVal, a practical framework that synergizes expert eye-tracking data with direct radiological evaluations to assess the quality of synthetic medical images. GazeVal leverages gaze patterns of radiologists as they provide a deeper understanding of how experts perceive and interact with synthetic data in different tasks (i.e., diagnostic or Turing tests). Experiments with sixteen radiologists revealed that 96.6% of the generated images (by the most recent state-of-the-art AI algorithm) were identified as fake, demonstrating the limitations of generative AI in producing clinically accurate images.
ScarNet: A Novel Foundation Model for Automated Myocardial Scar Quantification from LGE in Cardiac MRI
Jan 02, 2025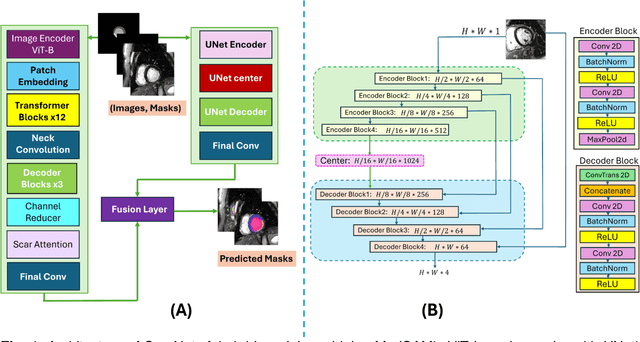
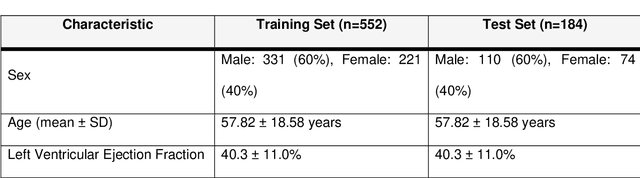
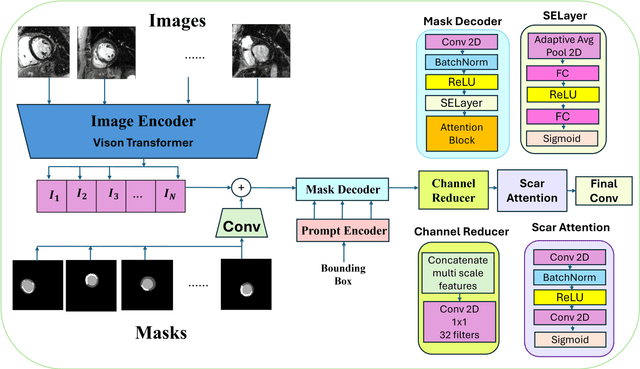
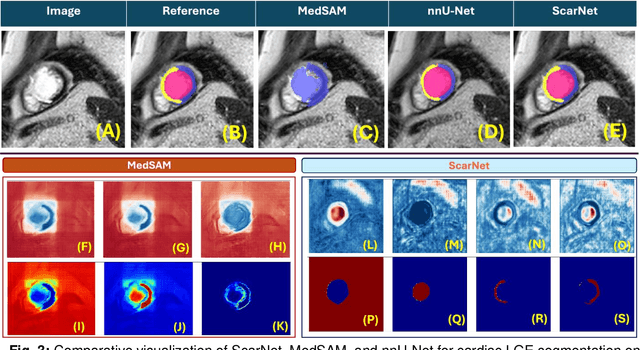
Abstract:Background: Late Gadolinium Enhancement (LGE) imaging is the gold standard for assessing myocardial fibrosis and scarring, with left ventricular (LV) LGE extent predicting major adverse cardiac events (MACE). Despite its importance, routine LGE-based LV scar quantification is hindered by labor-intensive manual segmentation and inter-observer variability. Methods: We propose ScarNet, a hybrid model combining a transformer-based encoder from the Medical Segment Anything Model (MedSAM) with a convolution-based U-Net decoder, enhanced by tailored attention blocks. ScarNet was trained on 552 ischemic cardiomyopathy patients with expert segmentations of myocardial and scar boundaries and tested on 184 separate patients. Results: ScarNet achieved robust scar segmentation in 184 test patients, yielding a median Dice score of 0.912 (IQR: 0.863--0.944), significantly outperforming MedSAM (median Dice = 0.046, IQR: 0.043--0.047) and nnU-Net (median Dice = 0.638, IQR: 0.604--0.661). ScarNet demonstrated lower bias (-0.63%) and coefficient of variation (4.3%) compared to MedSAM (bias: -13.31%, CoV: 130.3%) and nnU-Net (bias: -2.46%, CoV: 20.3%). In Monte Carlo simulations with noise perturbations, ScarNet achieved significantly higher scar Dice (0.892 \pm 0.053, CoV = 5.9%) than MedSAM (0.048 \pm 0.112, CoV = 233.3%) and nnU-Net (0.615 \pm 0.537, CoV = 28.7%). Conclusion: ScarNet outperformed MedSAM and nnU-Net in accurately segmenting myocardial and scar boundaries in LGE images. The model exhibited robust performance across diverse image qualities and scar patterns.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge