Neda Tavakoli
ScarNet: A Novel Foundation Model for Automated Myocardial Scar Quantification from LGE in Cardiac MRI
Jan 02, 2025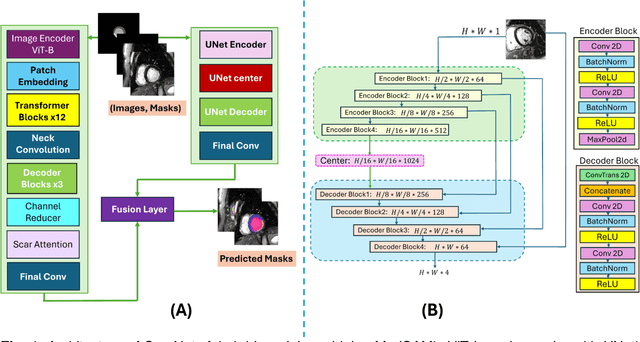
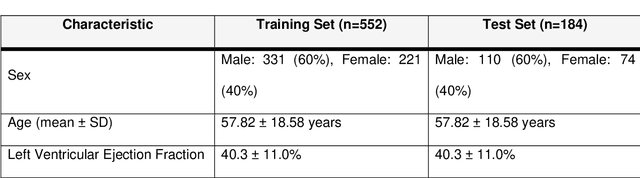
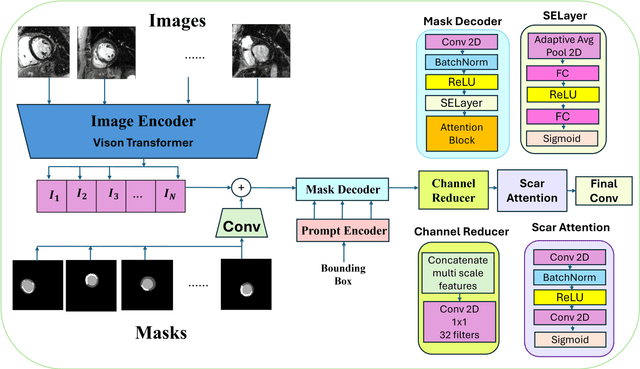
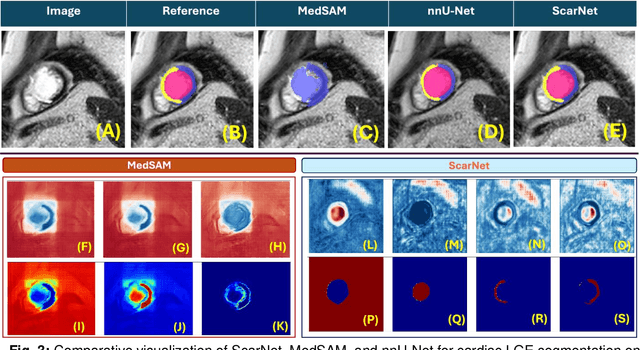
Abstract:Background: Late Gadolinium Enhancement (LGE) imaging is the gold standard for assessing myocardial fibrosis and scarring, with left ventricular (LV) LGE extent predicting major adverse cardiac events (MACE). Despite its importance, routine LGE-based LV scar quantification is hindered by labor-intensive manual segmentation and inter-observer variability. Methods: We propose ScarNet, a hybrid model combining a transformer-based encoder from the Medical Segment Anything Model (MedSAM) with a convolution-based U-Net decoder, enhanced by tailored attention blocks. ScarNet was trained on 552 ischemic cardiomyopathy patients with expert segmentations of myocardial and scar boundaries and tested on 184 separate patients. Results: ScarNet achieved robust scar segmentation in 184 test patients, yielding a median Dice score of 0.912 (IQR: 0.863--0.944), significantly outperforming MedSAM (median Dice = 0.046, IQR: 0.043--0.047) and nnU-Net (median Dice = 0.638, IQR: 0.604--0.661). ScarNet demonstrated lower bias (-0.63%) and coefficient of variation (4.3%) compared to MedSAM (bias: -13.31%, CoV: 130.3%) and nnU-Net (bias: -2.46%, CoV: 20.3%). In Monte Carlo simulations with noise perturbations, ScarNet achieved significantly higher scar Dice (0.892 \pm 0.053, CoV = 5.9%) than MedSAM (0.048 \pm 0.112, CoV = 233.3%) and nnU-Net (0.615 \pm 0.537, CoV = 28.7%). Conclusion: ScarNet outperformed MedSAM and nnU-Net in accurately segmenting myocardial and scar boundaries in LGE images. The model exhibited robust performance across diverse image qualities and scar patterns.
DRL-STNet: Unsupervised Domain Adaptation for Cross-modality Medical Image Segmentation via Disentangled Representation Learning
Sep 26, 2024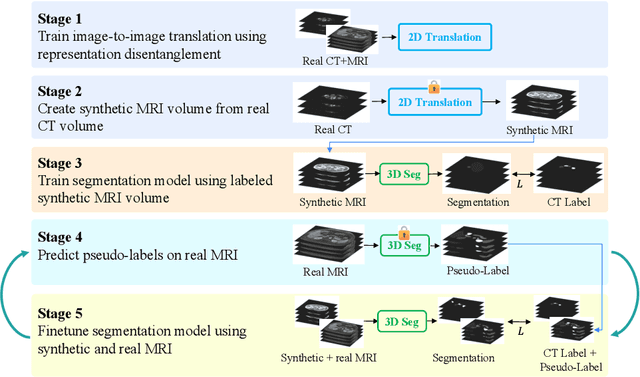

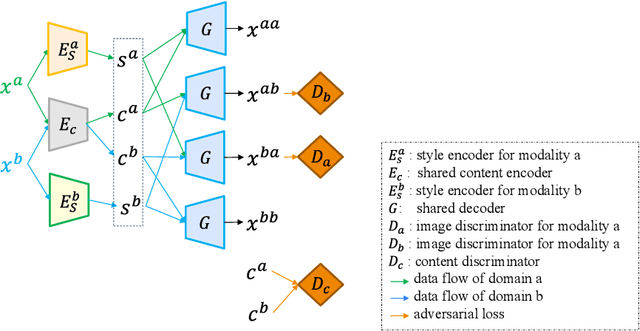
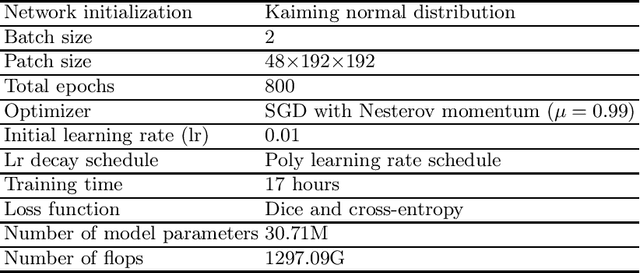
Abstract:Unsupervised domain adaptation (UDA) is essential for medical image segmentation, especially in cross-modality data scenarios. UDA aims to transfer knowledge from a labeled source domain to an unlabeled target domain, thereby reducing the dependency on extensive manual annotations. This paper presents DRL-STNet, a novel framework for cross-modality medical image segmentation that leverages generative adversarial networks (GANs), disentangled representation learning (DRL), and self-training (ST). Our method leverages DRL within a GAN to translate images from the source to the target modality. Then, the segmentation model is initially trained with these translated images and corresponding source labels and then fine-tuned iteratively using a combination of synthetic and real images with pseudo-labels and real labels. The proposed framework exhibits superior performance in abdominal organ segmentation on the FLARE challenge dataset, surpassing state-of-the-art methods by 11.4% in the Dice similarity coefficient and by 13.1% in the Normalized Surface Dice metric, achieving scores of 74.21% and 80.69%, respectively. The average running time is 41 seconds, and the area under the GPU memory-time curve is 11,292 MB. These results indicate the potential of DRL-STNet for enhancing cross-modality medical image segmentation tasks.
Fast Fourier Transformation for Optimizing Convolutional Neural Networks in Object Recognition
Oct 08, 2020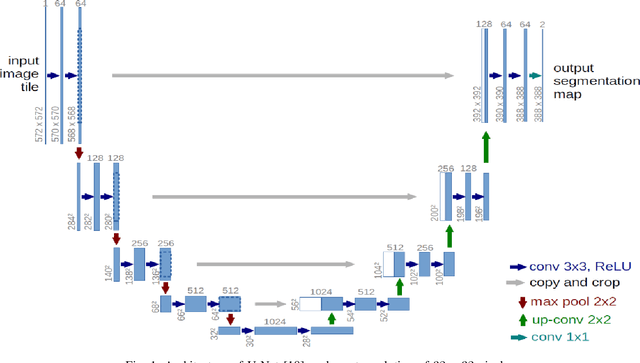
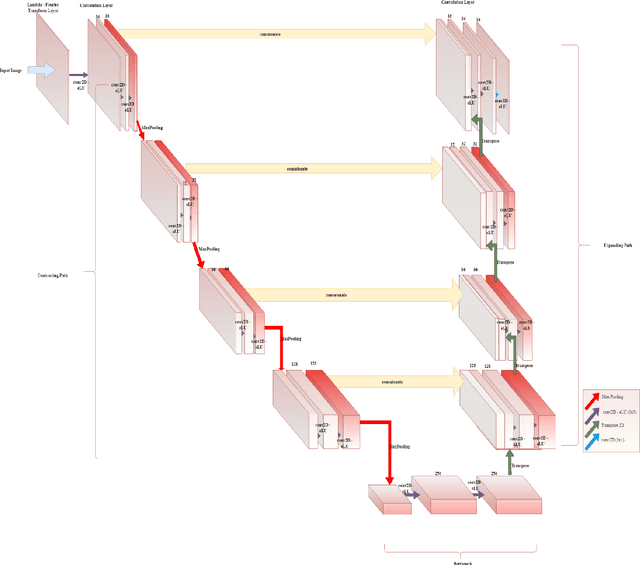
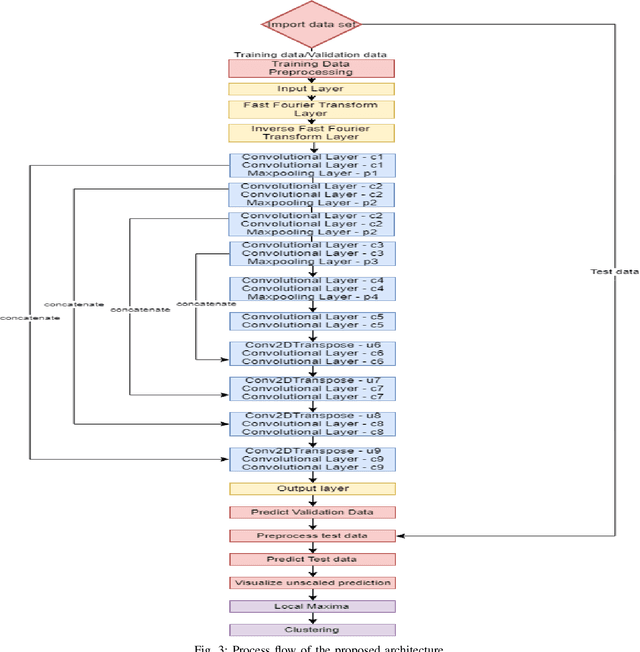
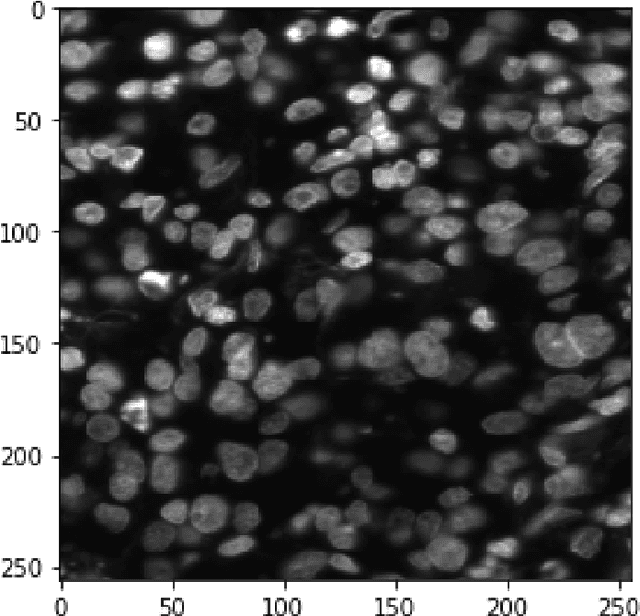
Abstract:This paper proposes to use Fast Fourier Transformation-based U-Net (a refined fully convolutional networks) and perform image convolution in neural networks. Leveraging the Fast Fourier Transformation, it reduces the image convolution costs involved in the Convolutional Neural Networks (CNNs) and thus reduces the overall computational costs. The proposed model identifies the object information from the images. We apply the Fast Fourier transform algorithm on an image data set to obtain more accessible information about the image data, before segmenting them through the U-Net architecture. More specifically, we implement the FFT-based convolutional neural network to improve the training time of the network. The proposed approach was applied to publicly available Broad Bioimage Benchmark Collection (BBBC) dataset. Our model demonstrated improvement in training time during convolution from $600-700$ ms/step to $400-500$ ms/step. We evaluated the accuracy of our model using Intersection over Union (IoU) metric showing significant improvements.
Clustering Time Series Data through Autoencoder-based Deep Learning Models
Apr 11, 2020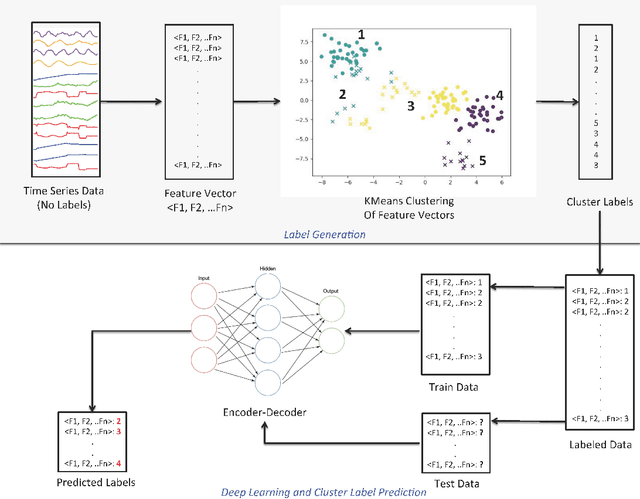
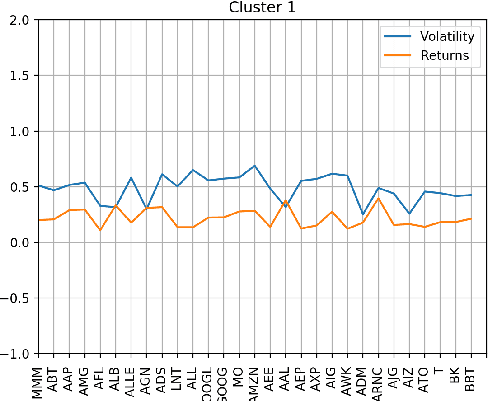
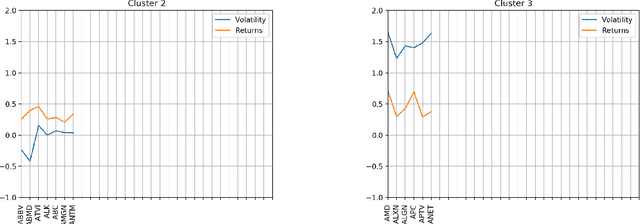
Abstract:Machine learning and in particular deep learning algorithms are the emerging approaches to data analysis. These techniques have transformed traditional data mining-based analysis radically into a learning-based model in which existing data sets along with their cluster labels (i.e., train set) are learned to build a supervised learning model and predict the cluster labels of unseen data (i.e., test set). In particular, deep learning techniques are capable of capturing and learning hidden features in a given data sets and thus building a more accurate prediction model for clustering and labeling problem. However, the major problem is that time series data are often unlabeled and thus supervised learning-based deep learning algorithms cannot be directly adapted to solve the clustering problems for these special and complex types of data sets. To address this problem, this paper introduces a two-stage method for clustering time series data. First, a novel technique is introduced to utilize the characteristics (e.g., volatility) of given time series data in order to create labels and thus be able to transform the problem from unsupervised learning into supervised learning. Second, an autoencoder-based deep learning model is built to learn and model both known and hidden features of time series data along with their created labels to predict the labels of unseen time series data. The paper reports a case study in which financial and stock time series data of selected 70 stock indices are clustered into distinct groups using the introduced two-stage procedure. The results show that the proposed procedure is capable of achieving 87.5\% accuracy in clustering and predicting the labels for unseen time series data.
Locality Sensitive Hashing-based Sequence Alignment Using Deep Bidirectional LSTM Models
Apr 05, 2020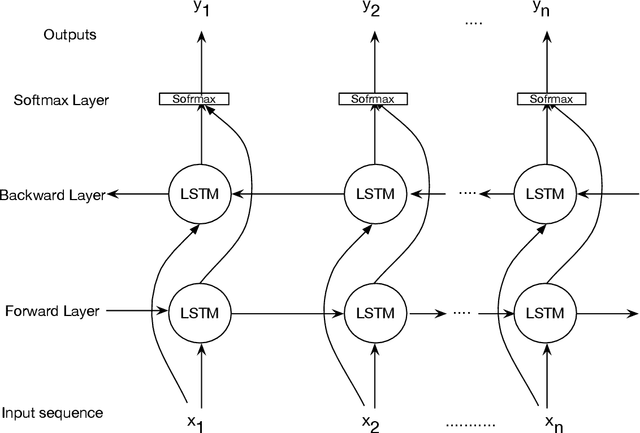
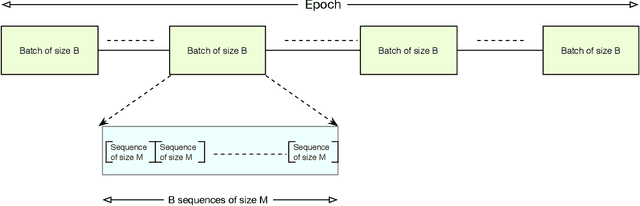
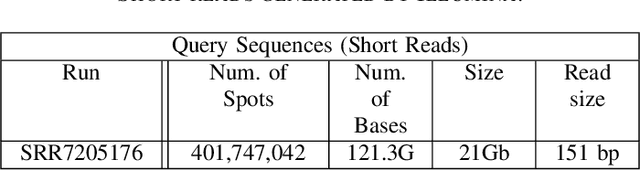
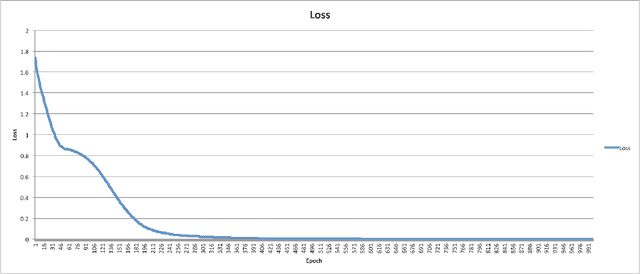
Abstract:Bidirectional Long Short-Term Memory (LSTM) is a special kind of Recurrent Neural Network (RNN) architecture which is designed to model sequences and their long-range dependencies more precisely than RNNs. This paper proposes to use deep bidirectional LSTM for sequence modeling as an approach to perform locality-sensitive hashing (LSH)-based sequence alignment. In particular, we use the deep bidirectional LSTM to learn features of LSH. The obtained LSH is then can be utilized to perform sequence alignment. We demonstrate the feasibility of the modeling sequences using the proposed LSTM-based model by aligning the short read queries over the reference genome. We use the human reference genome as our training dataset, in addition to a set of short reads generated using Illumina sequencing technology. The ultimate goal is to align query sequences into a reference genome. We first decompose the reference genome into multiple sequences. These sequences are then fed into the bidirectional LSTM model and then mapped into fixed-length vectors. These vectors are what we call the trained LSH, which can then be used for sequence alignment. The case study shows that using the introduced LSTM-based model, we achieve higher accuracy with the number of epochs.
A Comparative Analysis of Forecasting Financial Time Series Using ARIMA, LSTM, and BiLSTM
Nov 21, 2019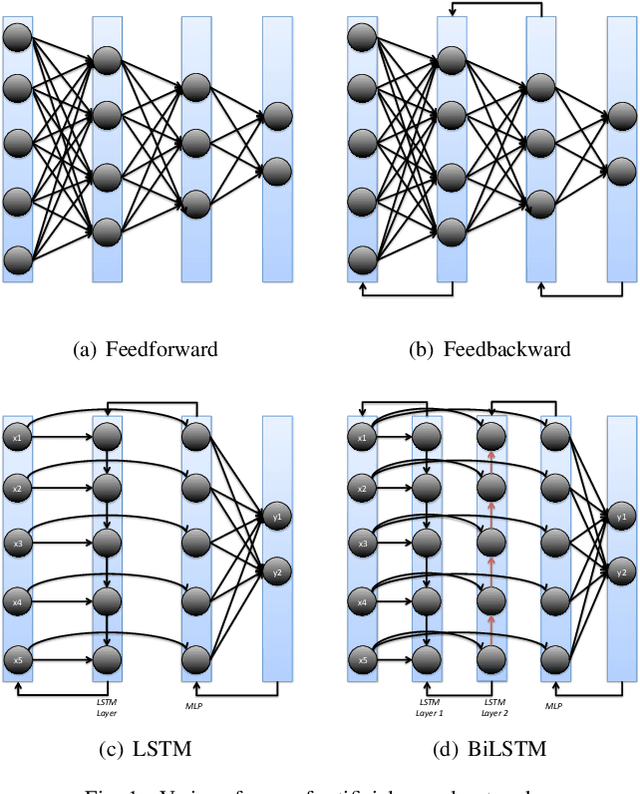
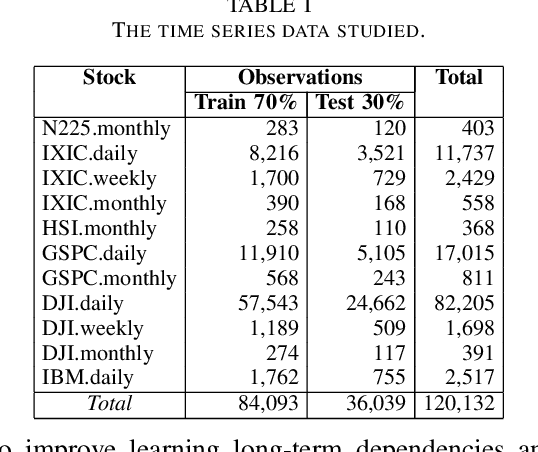
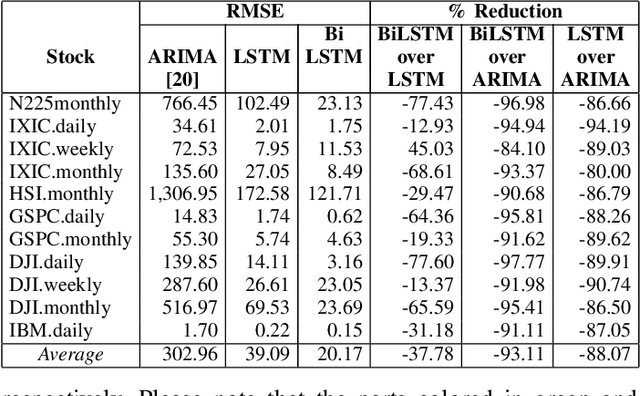
Abstract:Machine and deep learning-based algorithms are the emerging approaches in addressing prediction problems in time series. These techniques have been shown to produce more accurate results than conventional regression-based modeling. It has been reported that artificial Recurrent Neural Networks (RNN) with memory, such as Long Short-Term Memory (LSTM), are superior compared to Autoregressive Integrated Moving Average (ARIMA) with a large margin. The LSTM-based models incorporate additional "gates" for the purpose of memorizing longer sequences of input data. The major question is that whether the gates incorporated in the LSTM architecture already offers a good prediction and whether additional training of data would be necessary to further improve the prediction. Bidirectional LSTMs (BiLSTMs) enable additional training by traversing the input data twice (i.e., 1) left-to-right, and 2) right-to-left). The research question of interest is then whether BiLSTM, with additional training capability, outperforms regular unidirectional LSTM. This paper reports a behavioral analysis and comparison of BiLSTM and LSTM models. The objective is to explore to what extend additional layers of training of data would be beneficial to tune the involved parameters. The results show that additional training of data and thus BiLSTM-based modeling offers better predictions than regular LSTM-based models. More specifically, it was observed that BiLSTM models provide better predictions compared to ARIMA and LSTM models. It was also observed that BiLSTM models reach the equilibrium much slower than LSTM-based models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge