Amber L. Simpson
A Novel Patch-Based TDA Approach for Computed Tomography
Dec 13, 2025



Abstract:The development of machine learning (ML) models based on computed tomography (CT) imaging modality has been a major focus of recent research in the medical imaging domain. Incorporating robust feature engineering approach can highly improve the performance of these models. Topological data analysis (TDA), a recent development based on the mathematical field of algebraic topology, mainly focuses on the data from a topological perspective, extracting deeper insight and higher dimensional structures from the data. Persistent homology (PH), a fundamental tool in the area of TDA, can extract topological features such as connected components, cycles and voids from the data. A popular approach to construct PH from 3D CT images is to utilize the 3D cubical complex filtration, a method adapted for grid-structured data. However, this approach may not always yield the best performance and can suffer from computational complexity with higher resolution CT images. This study introduces a novel patch-based PH construction approach tailored for volumetric medical imaging data, in particular CT modality. A wide range of experiments has been conducted on several datasets of 3D CT images to comprehensively analyze the performance of the proposed method with various parameters and benchmark it against the 3D cubical complex algorithm. Our results highlight the dominance of the patch-based TDA approach in terms of both classification performance and time-efficiency. The proposed approach outperformed the cubical complex method, achieving average improvement of 10.38%, 6.94%, 2.06%, 11.58%, and 8.51% in accuracy, AUC, sensitivity, specificity, and F1 score, respectively, across all datasets. Finally, we provide a convenient python package, Patch-TDA, to facilitate the utilization of the proposed approach.
Comparing the Effects of Persistence Barcodes Aggregation and Feature Concatenation on Medical Imaging
May 29, 2025



Abstract:In medical image analysis, feature engineering plays an important role in the design and performance of machine learning models. Persistent homology (PH), from the field of topological data analysis (TDA), demonstrates robustness and stability to data perturbations and addresses the limitation from traditional feature extraction approaches where a small change in input results in a large change in feature representation. Using PH, we store persistent topological and geometrical features in the form of the persistence barcode whereby large bars represent global topological features and small bars encapsulate geometrical information of the data. When multiple barcodes are computed from 2D or 3D medical images, two approaches can be used to construct the final topological feature vector in each dimension: aggregating persistence barcodes followed by featurization or concatenating topological feature vectors derived from each barcode. In this study, we conduct a comprehensive analysis across diverse medical imaging datasets to compare the effects of the two aforementioned approaches on the performance of classification models. The results of this analysis indicate that feature concatenation preserves detailed topological information from individual barcodes, yields better classification performance and is therefore a preferred approach when conducting similar experiments.
Finding Reproducible and Prognostic Radiomic Features in Variable Slice Thickness Contrast Enhanced CT of Colorectal Liver Metastases
Jan 20, 2025



Abstract:Establishing the reproducibility of radiomic signatures is a critical step in the path to clinical adoption of quantitative imaging biomarkers; however, radiomic signatures must also be meaningfully related to an outcome of clinical importance to be of value for personalized medicine. In this study, we analyze both the reproducibility and prognostic value of radiomic features extracted from the liver parenchyma and largest liver metastases in contrast enhanced CT scans of patients with colorectal liver metastases (CRLM). A prospective cohort of 81 patients from two major US cancer centers was used to establish the reproducibility of radiomic features extracted from images reconstructed with different slice thicknesses. A publicly available, single-center cohort of 197 preoperative scans from patients who underwent hepatic resection for treatment of CRLM was used to evaluate the prognostic value of features and models to predict overall survival. A standard set of 93 features was extracted from all images, with a set of eight different extractor settings. The feature extraction settings producing the most reproducible, as well as the most prognostically discriminative feature values were highly dependent on both the region of interest and the specific feature in question. While the best overall predictive model was produced using features extracted with a particular setting, without accounting for reproducibility, (C-index = 0.630 (0.603--0.649)) an equivalent-performing model (C-index = 0.629 (0.605--0.645)) was produced by pooling features from all extraction settings, and thresholding features with low reproducibility ($\mathrm{CCC} \geq 0.85$), prior to feature selection. Our findings support a data-driven approach to feature extraction and selection, preferring the inclusion of many features, and narrowing feature selection based on reproducibility when relevant data is available.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024:032
Parameter-Efficient Methods for Metastases Detection from Clinical Notes
Oct 27, 2023

Abstract:Understanding the progression of cancer is crucial for defining treatments for patients. The objective of this study is to automate the detection of metastatic liver disease from free-style computed tomography (CT) radiology reports. Our research demonstrates that transferring knowledge using three approaches can improve model performance. First, we utilize generic language models (LMs), pretrained in a self-supervised manner. Second, we use a semi-supervised approach to train our model by automatically annotating a large unlabeled dataset; this approach substantially enhances the model's performance. Finally, we transfer knowledge from related tasks by designing a multi-task transfer learning methodology. We leverage the recent advancement of parameter-efficient LM adaptation strategies to improve performance and training efficiency. Our dataset consists of CT reports collected at Memorial Sloan Kettering Cancer Center (MSKCC) over the course of 12 years. 2,641 reports were manually annotated by domain experts; among them, 841 reports have been annotated for the presence of liver metastases. Our best model achieved an F1-score of 73.8%, a precision of 84%, and a recall of 65.8%.
* 6 pages, 1 figure, The 36th Canadian Conference on Artificial Intelligence
Towards Optimal Patch Size in Vision Transformers for Tumor Segmentation
Aug 31, 2023Abstract:Detection of tumors in metastatic colorectal cancer (mCRC) plays an essential role in the early diagnosis and treatment of liver cancer. Deep learning models backboned by fully convolutional neural networks (FCNNs) have become the dominant model for segmenting 3D computerized tomography (CT) scans. However, since their convolution layers suffer from limited kernel size, they are not able to capture long-range dependencies and global context. To tackle this restriction, vision transformers have been introduced to solve FCNN's locality of receptive fields. Although transformers can capture long-range features, their segmentation performance decreases with various tumor sizes due to the model sensitivity to the input patch size. While finding an optimal patch size improves the performance of vision transformer-based models on segmentation tasks, it is a time-consuming and challenging procedure. This paper proposes a technique to select the vision transformer's optimal input multi-resolution image patch size based on the average volume size of metastasis lesions. We further validated our suggested framework using a transfer-learning technique, demonstrating that the highest Dice similarity coefficient (DSC) performance was obtained by pre-training on training data with a larger tumour volume using the suggested ideal patch size and then training with a smaller one. We experimentally evaluate this idea through pre-training our model on a multi-resolution public dataset. Our model showed consistent and improved results when applied to our private multi-resolution mCRC dataset with a smaller average tumor volume. This study lays the groundwork for optimizing semantic segmentation of small objects using vision transformers. The implementation source code is available at:https://github.com/Ramtin-Mojtahedi/OVTPS.
Attention-based CT Scan Interpolation for Lesion Segmentation of Colorectal Liver Metastases
Aug 30, 2023Abstract:Small liver lesions common to colorectal liver metastases (CRLMs) are challenging for convolutional neural network (CNN) segmentation models, especially when we have a wide range of slice thicknesses in the computed tomography (CT) scans. Slice thickness of CT images may vary by clinical indication. For example, thinner slices are used for presurgical planning when fine anatomic details of small vessels are required. While keeping the effective radiation dose in patients as low as possible, various slice thicknesses are employed in CRLMs due to their limitations. However, differences in slice thickness across CTs lead to significant performance degradation in CT segmentation models based on CNNs. This paper proposes a novel unsupervised attention-based interpolation model to generate intermediate slices from consecutive triplet slices in CT scans. We integrate segmentation loss during the interpolation model's training to leverage segmentation labels in existing slices to generate middle ones. Unlike common interpolation techniques in CT volumes, our model highlights the regions of interest (liver and lesions) inside the abdominal CT scans in the interpolated slice. Moreover, our model's outputs are consistent with the original input slices while increasing the segmentation performance in two cutting-edge 3D segmentation pipelines. We tested the proposed model on the CRLM dataset to upsample subjects with thick slices and create isotropic volume for our segmentation model. The produced isotropic dataset increases the Dice score in the segmentation of lesions and outperforms other interpolation approaches in terms of interpolation metrics.
Modality Completion via Gaussian Process Prior Variational Autoencoders for Multi-Modal Glioma Segmentation
Jul 07, 2021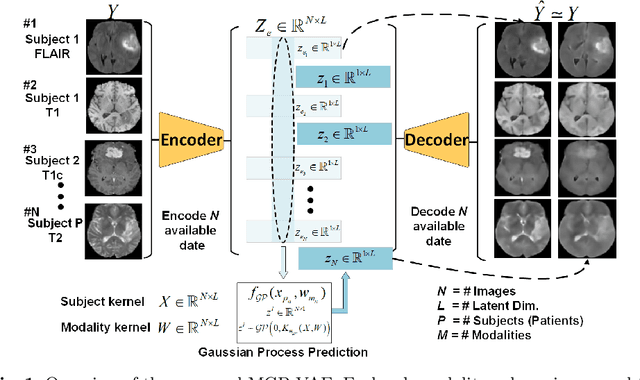
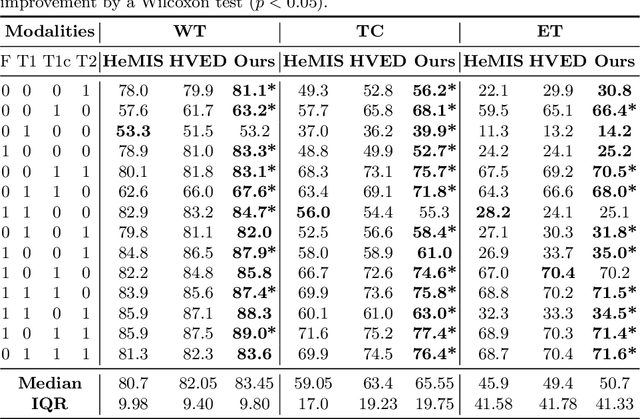
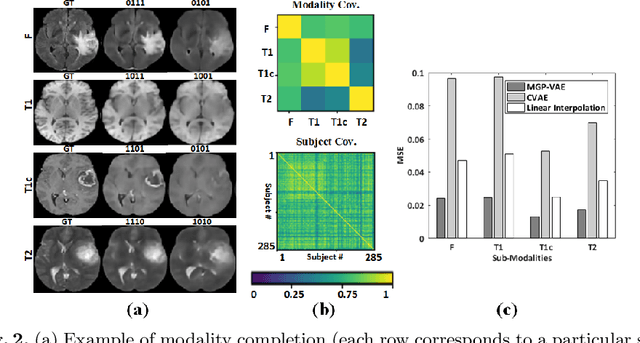
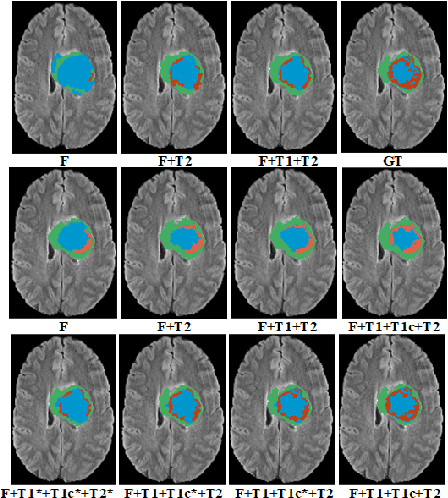
Abstract:In large studies involving multi protocol Magnetic Resonance Imaging (MRI), it can occur to miss one or more sub-modalities for a given patient owing to poor quality (e.g. imaging artifacts), failed acquisitions, or hallway interrupted imaging examinations. In some cases, certain protocols are unavailable due to limited scan time or to retrospectively harmonise the imaging protocols of two independent studies. Missing image modalities pose a challenge to segmentation frameworks as complementary information contributed by the missing scans is then lost. In this paper, we propose a novel model, Multi-modal Gaussian Process Prior Variational Autoencoder (MGP-VAE), to impute one or more missing sub-modalities for a patient scan. MGP-VAE can leverage the Gaussian Process (GP) prior on the Variational Autoencoder (VAE) to utilize the subjects/patients and sub-modalities correlations. Instead of designing one network for each possible subset of present sub-modalities or using frameworks to mix feature maps, missing data can be generated from a single model based on all the available samples. We show the applicability of MGP-VAE on brain tumor segmentation where either, two, or three of four sub-modalities may be missing. Our experiments against competitive segmentation baselines with missing sub-modality on BraTS'19 dataset indicate the effectiveness of the MGP-VAE model for segmentation tasks.
The Medical Segmentation Decathlon
Jun 10, 2021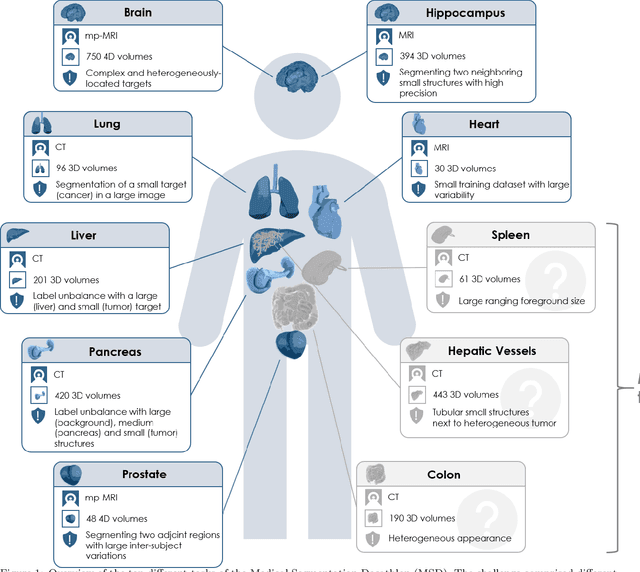
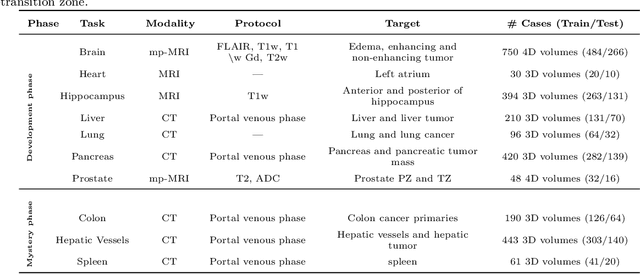
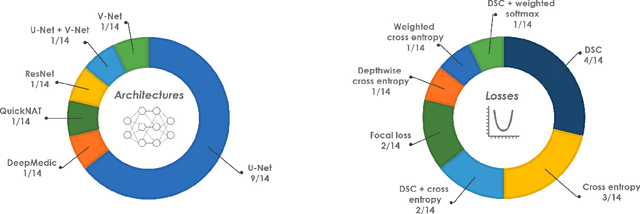
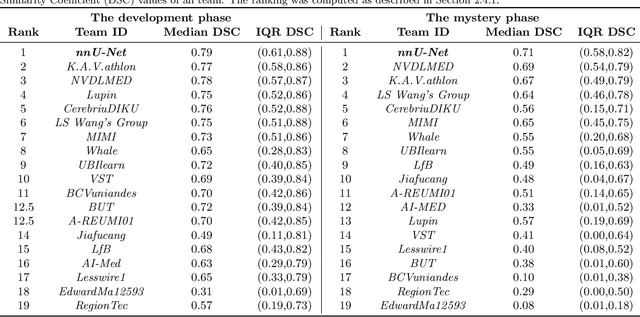
Abstract:International challenges have become the de facto standard for comparative assessment of image analysis algorithms given a specific task. Segmentation is so far the most widely investigated medical image processing task, but the various segmentation challenges have typically been organized in isolation, such that algorithm development was driven by the need to tackle a single specific clinical problem. We hypothesized that a method capable of performing well on multiple tasks will generalize well to a previously unseen task and potentially outperform a custom-designed solution. To investigate the hypothesis, we organized the Medical Segmentation Decathlon (MSD) - a biomedical image analysis challenge, in which algorithms compete in a multitude of both tasks and modalities. The underlying data set was designed to explore the axis of difficulties typically encountered when dealing with medical images, such as small data sets, unbalanced labels, multi-site data and small objects. The MSD challenge confirmed that algorithms with a consistent good performance on a set of tasks preserved their good average performance on a different set of previously unseen tasks. Moreover, by monitoring the MSD winner for two years, we found that this algorithm continued generalizing well to a wide range of other clinical problems, further confirming our hypothesis. Three main conclusions can be drawn from this study: (1) state-of-the-art image segmentation algorithms are mature, accurate, and generalize well when retrained on unseen tasks; (2) consistent algorithmic performance across multiple tasks is a strong surrogate of algorithmic generalizability; (3) the training of accurate AI segmentation models is now commoditized to non AI experts.
A large annotated medical image dataset for the development and evaluation of segmentation algorithms
Feb 25, 2019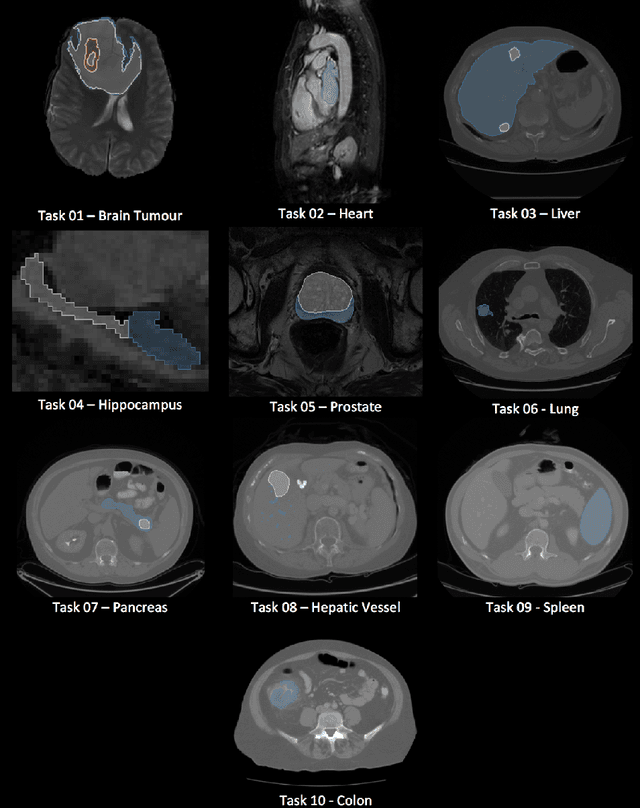

Abstract:Semantic segmentation of medical images aims to associate a pixel with a label in a medical image without human initialization. The success of semantic segmentation algorithms is contingent on the availability of high-quality imaging data with corresponding labels provided by experts. We sought to create a large collection of annotated medical image datasets of various clinically relevant anatomies available under open source license to facilitate the development of semantic segmentation algorithms. Such a resource would allow: 1) objective assessment of general-purpose segmentation methods through comprehensive benchmarking and 2) open and free access to medical image data for any researcher interested in the problem domain. Through a multi-institutional effort, we generated a large, curated dataset representative of several highly variable segmentation tasks that was used in a crowd-sourced challenge - the Medical Segmentation Decathlon held during the 2018 Medical Image Computing and Computer Aided Interventions Conference in Granada, Spain. Here, we describe these ten labeled image datasets so that these data may be effectively reused by the research community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge