Alexandre X. Falcão
Co-Training with Active Contrastive Learning and Meta-Pseudo-Labeling on 2D Projections for Deep Semi-Supervised Learning
Apr 25, 2025Abstract:A major challenge that prevents the training of DL models is the limited availability of accurately labeled data. This shortcoming is highlighted in areas where data annotation becomes a time-consuming and error-prone task. In this regard, SSL tackles this challenge by capitalizing on scarce labeled and abundant unlabeled data; however, SoTA methods typically depend on pre-trained features and large validation sets to learn effective representations for classification tasks. In addition, the reduced set of labeled data is often randomly sampled, neglecting the selection of more informative samples. Here, we present active-DeepFA, a method that effectively combines CL, teacher-student-based meta-pseudo-labeling and AL to train non-pretrained CNN architectures for image classification in scenarios of scarcity of labeled and abundance of unlabeled data. It integrates DeepFA into a co-training setup that implements two cooperative networks to mitigate confirmation bias from pseudo-labels. The method starts with a reduced set of labeled samples by warming up the networks with supervised CL. Afterward and at regular epoch intervals, label propagation is performed on the 2D projections of the networks' deep features. Next, the most reliable pseudo-labels are exchanged between networks in a cross-training fashion, while the most meaningful samples are annotated and added into the labeled set. The networks independently minimize an objective loss function comprising supervised contrastive, supervised and semi-supervised loss components, enhancing the representations towards image classification. Our approach is evaluated on three challenging biological image datasets using only 5% of labeled samples, improving baselines and outperforming six other SoTA methods. In addition, it reduces annotation effort by achieving comparable results to those of its counterparts with only 3% of labeled data.
Multi-level Cellular Automata for FLIM networks
Apr 15, 2025Abstract:The necessity of abundant annotated data and complex network architectures presents a significant challenge in deep-learning Salient Object Detection (deep SOD) and across the broader deep-learning landscape. This challenge is particularly acute in medical applications in developing countries with limited computational resources. Combining modern and classical techniques offers a path to maintaining competitive performance while enabling practical applications. Feature Learning from Image Markers (FLIM) methodology empowers experts to design convolutional encoders through user-drawn markers, with filters learned directly from these annotations. Recent findings demonstrate that coupling a FLIM encoder with an adaptive decoder creates a flyweight network suitable for SOD, requiring significantly fewer parameters than lightweight models and eliminating the need for backpropagation. Cellular Automata (CA) methods have proven successful in data-scarce scenarios but require proper initialization -- typically through user input, priors, or randomness. We propose a practical intersection of these approaches: using FLIM networks to initialize CA states with expert knowledge without requiring user interaction for each image. By decoding features from each level of a FLIM network, we can initialize multiple CAs simultaneously, creating a multi-level framework. Our method leverages the hierarchical knowledge encoded across different network layers, merging multiple saliency maps into a high-quality final output that functions as a CA ensemble. Benchmarks across two challenging medical datasets demonstrate the competitiveness of our multi-level CA approach compared to established models in the deep SOD literature.
Interactive Image Selection and Training for Brain Tumor Segmentation Network
Jun 05, 2024Abstract:Medical image segmentation is a relevant problem, with deep learning being an exponent. However, the necessity of a high volume of fully annotated images for training massive models can be a problem, especially for applications whose images present a great diversity, such as brain tumors, which can occur in different sizes and shapes. In contrast, a recent methodology, Feature Learning from Image Markers (FLIM), has involved an expert in the learning loop, producing small networks that require few images to train the convolutional layers. In this work, We employ an interactive method for image selection and training based on FLIM, exploring the user's knowledge. The results demonstrated that with our methodology, we could choose a small set of images to train the encoder of a U-shaped network, obtaining performance equal to manual selection and even surpassing the same U-shaped network trained with backpropagation and all training images.
Building Brain Tumor Segmentation Networks with User-Assisted Filter Estimation and Selection
Mar 19, 2024Abstract:Brain tumor image segmentation is a challenging research topic in which deep-learning models have presented the best results. However, the traditional way of training those models from many pre-annotated images leaves several unanswered questions. Hence methodologies, such as Feature Learning from Image Markers (FLIM), have involved an expert in the learning loop to reduce human effort in data annotation and build models sufficiently deep for a given problem. FLIM has been successfully used to create encoders, estimating the filters of all convolutional layers from patches centered at marker voxels. In this work, we present Multi-Step (MS) FLIM - a user-assisted approach to estimating and selecting the most relevant filters from multiple FLIM executions. MS-FLIM is used only for the first convolutional layer, and the results already indicate improvement over FLIM. For evaluation, we build a simple U-shaped encoder-decoder network, named sU-Net, for glioblastoma segmentation using T1Gd and FLAIR MRI scans, varying the encoder's training method, using FLIM, MS-FLIM, and backpropagation algorithm. Also, we compared these sU-Nets with two State-Of-The-Art (SOTA) deep-learning models using two datasets. The results show that the sU-Net based on MS-FLIM outperforms the other training methods and achieves effectiveness within the standard deviations of the SOTA models.
CNN Filter Learning from Drawn Markers for the Detection of Suggestive Signs of COVID-19 in CT Images
Nov 16, 2021



Abstract:Early detection of COVID-19 is vital to control its spread. Deep learning methods have been presented to detect suggestive signs of COVID-19 from chest CT images. However, due to the novelty of the disease, annotated volumetric data are scarce. Here we propose a method that does not require either large annotated datasets or backpropagation to estimate the filters of a convolutional neural network (CNN). For a few CT images, the user draws markers at representative normal and abnormal regions. The method generates a feature extractor composed of a sequence of convolutional layers, whose kernels are specialized in enhancing regions similar to the marked ones, and the decision layer of our CNN is a support vector machine. As we have no control over the CT image acquisition, we also propose an intensity standardization approach. Our method can achieve mean accuracy and kappa values of $0.97$ and $0.93$, respectively, on a dataset with 117 CT images extracted from different sites, surpassing its counterpart in all scenarios.
Hierarchical Learning Using Deep Optimum-Path Forest
Feb 18, 2021



Abstract:Bag-of-Visual Words (BoVW) and deep learning techniques have been widely used in several domains, which include computer-assisted medical diagnoses. In this work, we are interested in developing tools for the automatic identification of Parkinson's disease using machine learning and the concept of BoVW. The proposed approach concerns a hierarchical-based learning technique to design visual dictionaries through the Deep Optimum-Path Forest classifier. The proposed method was evaluated in six datasets derived from data collected from individuals when performing handwriting exams. Experimental results showed the potential of the technique, with robust achievements.
Convolutional Neural Networks from Image Markers
Dec 15, 2020


Abstract:A technique named Feature Learning from Image Markers (FLIM) was recently proposed to estimate convolutional filters, with no backpropagation, from strokes drawn by a user on very few images (e.g., 1-3) per class, and demonstrated for coconut-tree image classification. This paper extends FLIM for fully connected layers and demonstrates it on different image classification problems. The work evaluates marker selection from multiple users and the impact of adding a fully connected layer. The results show that FLIM-based convolutional neural networks can outperform the same architecture trained from scratch by backpropagation.
Deploying machine learning to assist digital humanitarians: making image annotation in OpenStreetMap more efficient
Sep 17, 2020



Abstract:Locating populations in rural areas of developing countries has attracted the attention of humanitarian mapping projects since it is important to plan actions that affect vulnerable areas. Recent efforts have tackled this problem as the detection of buildings in aerial images. However, the quality and the amount of rural building annotated data in open mapping services like OpenStreetMap (OSM) is not sufficient for training accurate models for such detection. Although these methods have the potential of aiding in the update of rural building information, they are not accurate enough to automatically update the rural building maps. In this paper, we explore a human-computer interaction approach and propose an interactive method to support and optimize the work of volunteers in OSM. The user is asked to verify/correct the annotation of selected tiles during several iterations and therefore improving the model with the new annotated data. The experimental results, with simulated and real user annotation corrections, show that the proposed method greatly reduces the amount of data that the volunteers of OSM need to verify/correct. The proposed methodology could benefit humanitarian mapping projects, not only by making more efficient the process of annotation but also by improving the engagement of volunteers.
Correcting rural building annotations in OpenStreetMap using convolutional neural networks
Jan 24, 2019



Abstract:Rural building mapping is paramount to support demographic studies and plan actions in response to crisis that affect those areas. Rural building annotations exist in OpenStreetMap (OSM), but their quality and quantity are not sufficient for training models that can create accurate rural building maps. The problems with these annotations essentially fall into three categories: (i) most commonly, many annotations are geometrically misaligned with the updated imagery; (ii) some annotations do not correspond to buildings in the images (they are misannotations or the buildings have been destroyed); and (iii) some annotations are missing for buildings in the images (the buildings were never annotated or were built between subsequent image acquisitions). First, we propose a method based on Markov Random Field (MRF) to align the buildings with their annotations. The method maximizes the correlation between annotations and a building probability map while enforcing that nearby buildings have similar alignment vectors. Second, the annotations with no evidence in the building probability map are removed. Third, we present a method to detect non-annotated buildings with predefined shapes and add their annotation. The proposed methodology shows considerable improvement in accuracy of the OSM annotations for two regions of Tanzania and Zimbabwe, being more accurate than state-of-the-art baselines.
An Iterative Spanning Forest Framework for Superpixel Segmentation
Jan 30, 2018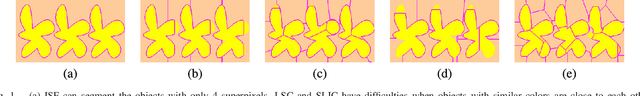
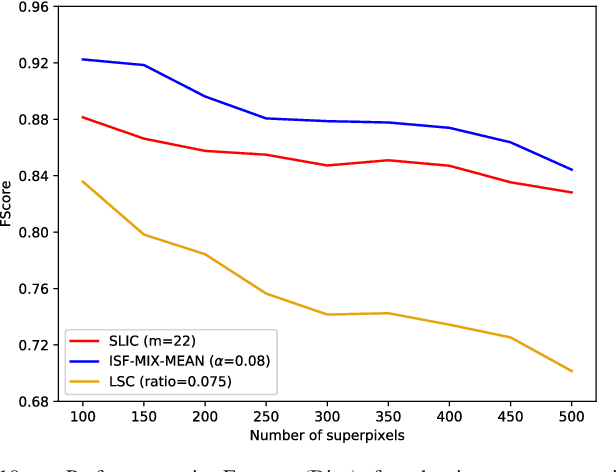
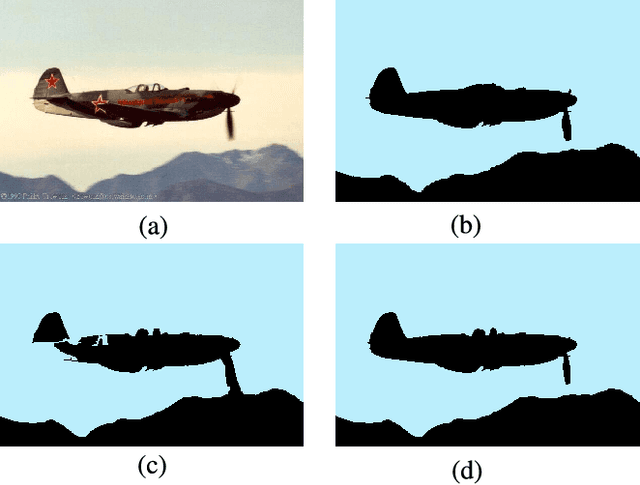

Abstract:Superpixel segmentation has become an important research problem in image processing. In this paper, we propose an Iterative Spanning Forest (ISF) framework, based on sequences of Image Foresting Transforms, where one can choose i) a seed sampling strategy, ii) a connectivity function, iii) an adjacency relation, and iv) a seed pixel recomputation procedure to generate improved sets of connected superpixels (supervoxels in 3D) per iteration. The superpixels in ISF structurally correspond to spanning trees rooted at those seeds. We present five ISF methods to illustrate different choices of its components. These methods are compared with approaches from the state-of-the-art in effectiveness and efficiency. The experiments involve 2D and 3D datasets with distinct characteristics, and a high level application, named sky image segmentation. The theoretical properties of ISF are demonstrated in the supplementary material and the results show that some of its methods are competitive with or superior to the best baselines in effectiveness and efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge