Alexandre Cunha
Deep Metric Structured Learning For Facial Expression Recognition
Jan 18, 2020



Abstract:We propose a deep metric learning model to create embedded sub-spaces with a well defined structure. A new loss function that imposes Gaussian structures on the output space is introduced to create these sub-spaces thus shaping the distribution of the data. Having a mixture of Gaussians solution space is advantageous given its simplified and well established structure. It allows fast discovering of classes within classes and the identification of mean representatives at the centroids of individual classes. We also propose a new semi-supervised method to create sub-classes. We illustrate our methods on the facial expression recognition problem and validate results on the FER+, AffectNet, Extended Cohn-Kanade (CK+), BU-3DFE, and JAFFE datasets. We experimentally demonstrate that the learned embedding can be successfully used for various applications including expression retrieval and emotion recognition.
J Regularization Improves Imbalanced Multiclass Segmentation
Oct 22, 2019



Abstract:We propose a new loss formulation to further advance the multiclass segmentation of cluttered cells under weakly supervised conditions. We improve the separation of touching and immediate cells, obtaining sharp segmentation boundaries with high adequacy, when we add Youden's $J$ statistic regularization term to the cross entropy loss. This regularization intrinsically supports class imbalance thus eliminating the necessity of explicitly using weights to balance training. Simulations demonstrate this capability and show how the regularization leads to better results by helping advancing the optimization when cross entropy stalls. We build upon our previous work on multiclass segmentation by adding yet another training class representing gaps between adjacent cells. This addition helps the classifier identify narrow gaps as background and no longer as touching regions. We present results of our methods for 2D and 3D images, from bright field to confocal stacks containing different types of cells, and we show that they accurately segment individual cells after training with a limited number of annotated images, some of which are poorly annotated.
A Multiple Source Hourglass Deep Network for Multi-Focus Image Fusion
Aug 28, 2019


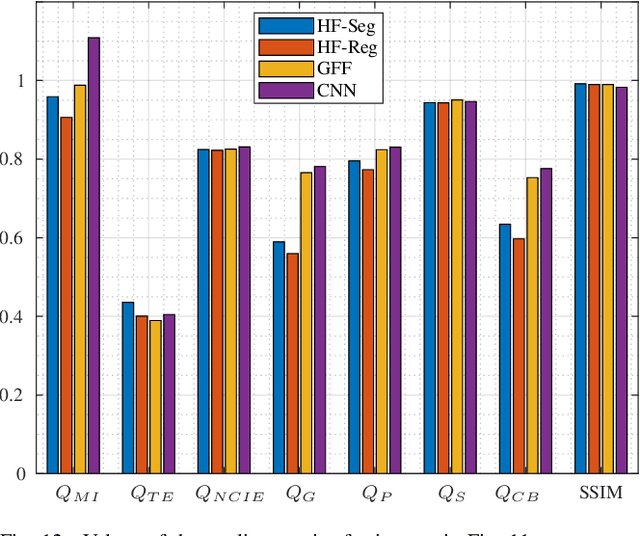
Abstract:Multi-Focus Image Fusion seeks to improve the quality of an acquired burst of images with different focus planes. For solving the task, an activity level measurement and a fusion rule are typically established to select and fuse the most relevant information from the sources. However, the design of this kind of method by hand is really hard and sometimes restricted to solution spaces where the optimal all-in-focus images are not contained. Then, we propose here two fast and straightforward approaches for image fusion based on deep neural networks. Our solution uses a multiple source Hourglass architecture trained in an end-to-end fashion. Models are data-driven and can be easily generalized for other kinds of fusion problems. A segmentation approach is used for recognition of the focus map, while the weighted average rule is used for fusion. We designed a training loss function for our regression-based fusion function, which allows the network to learn both the activity level measurement and the fusion rule. Experimental results show our approach has comparable results to the state-of-the-art methods with a 60X increase of computational efficiency for 520X520 resolution images.
A Weakly Supervised Method for Instance Segmentation of Biological Cells
Aug 26, 2019



Abstract:We present a weakly supervised deep learning method to perform instance segmentation of cells present in microscopy images. Annotation of biomedical images in the lab can be scarce, incomplete, and inaccurate. This is of concern when supervised learning is used for image analysis as the discriminative power of a learning model might be compromised in these situations. To overcome the curse of poor labeling, our method focuses on three aspects to improve learning: i) we propose a loss function operating in three classes to facilitate separating adjacent cells and to drive the optimizer to properly classify underrepresented regions; ii) a contour-aware weight map model is introduced to strengthen contour detection while improving the network generalization capacity; and iii) we augment data by carefully modulating local intensities on edges shared by adjoining regions and to account for possibly weak signals on these edges. Generated probability maps are segmented using different methods, with the watershed based one generally offering the best solutions, specially in those regions where the prevalence of a single class is not clear. The combination of these contributions allows segmenting individual cells on challenging images. We demonstrate our methods in sparse and crowded cell images, showing improvements in the learning process for a fixed network architecture.
FERAtt: Facial Expression Recognition with Attention Net
Feb 08, 2019



Abstract:We present a new end-to-end network architecture for facial expression recognition with an attention model. It focuses attention in the human face and uses a Gaussian space representation for expression recognition. We devise this architecture based on two fundamental complementary components: (1) facial image correction and attention and (2) facial expression representation and classification. The first component uses an encoder-decoder style network and a convolutional feature extractor that are pixel-wise multiplied to obtain a feature attention map. The second component is responsible for obtaining an embedded representation and classification of the facial expression. We propose a loss function that creates a Gaussian structure on the representation space. To demonstrate the proposed method, we create two larger and more comprehensive synthetic datasets using the traditional BU3DFE and CK+ facial datasets. We compared results with the PreActResNet18 baseline. Our experiments on these datasets have shown the superiority of our approach in recognizing facial expressions.
Geometric Median Shapes
Oct 29, 2018



Abstract:We present an algorithm to compute the geometric median of shapes which is based on the extension of median to high dimensions. The median finding problem is formulated as an optimization over distances and it is solved directly using the watershed method as an optimizer. We show that computing the geometric median of shapes is robust in the presence of outliers and it is superior to the mean shape which can easily be affected by the presence of outliers. The geometric median shape thus faithfully represents the true central tendency of the data, contaminated or not. Our approach can be applied to manifold and non manifold shapes, with connected or disconnected shapes. The application of distance transforms and watershed algorithm, two well established constructs of image processing, lead to an algorithm that can be quickly implemented to generate fast solutions with linear storage requirements. We demonstrate our methods in synthetic and natural shapes and compare median and mean results under increasing contamination by strong outliers.
CNN-based Preprocessing to Optimize Watershed-based Cell Segmentation in 3D Confocal Microscopy Images
Oct 16, 2018



Abstract:The quantitative analysis of cellular membranes helps understanding developmental processes at the cellular level. Particularly 3D microscopic image data offers valuable insights into cell dynamics, but error-free automatic segmentation remains challenging due to the huge amount of data generated and strong variations in image intensities. In this paper, we propose a new 3D segmentation approach which combines the discriminative power of convolutional neural networks (CNNs) for preprocessing and investigates the performance of three watershed-based postprocessing strategies (WS), which are well suited to segment object shapes, even when supplied with vague seed and boundary constraints. To leverage the full potential of the watershed algorithm, the multi-instance segmentation problem is initially interpreted as three-class semantic segmentation problem, which in turn is well-suited for the application of CNNs. Using manually annotated 3D confocal microscopy images of Arabidopsis thaliana, we show the superior performance of the proposed method compared to the state of the art.
Multiclass Weighted Loss for Instance Segmentation of Cluttered Cells
Feb 21, 2018
Abstract:We propose a new multiclass weighted loss function for instance segmentation of cluttered cells. We are primarily motivated by the need of developmental biologists to quantify and model the behavior of blood T-cells which might help us in understanding their regulation mechanisms and ultimately help researchers in their quest for developing an effective immuno-therapy cancer treatment. Segmenting individual touching cells in cluttered regions is challenging as the feature distribution on shared borders and cell foreground are similar thus difficulting discriminating pixels into proper classes. We present two novel weight maps applied to the weighted cross entropy loss function which take into account both class imbalance and cell geometry. Binary ground truth training data is augmented so the learning model can handle not only foreground and background but also a third touching class. This framework allows training using U-Net. Experiments with our formulations have shown superior results when compared to other similar schemes, outperforming binary class models with significant improvement of boundary adequacy and instance detection. We validate our results on manually annotated microscope images of T-cells.
SEGMENT3D: A Web-based Application for Collaborative Segmentation of 3D images used in the Shoot Apical Meristem
Oct 26, 2017



Abstract:The quantitative analysis of 3D confocal microscopy images of the shoot apical meristem helps understanding the growth process of some plants. Cell segmentation in these images is crucial for computational plant analysis and many automated methods have been proposed. However, variations in signal intensity across the image mitigate the effectiveness of those approaches with no easy way for user correction. We propose a web-based collaborative 3D image segmentation application, SEGMENT3D, to leverage automatic segmentation results. The image is divided into 3D tiles that can be either segmented interactively from scratch or corrected from a pre-existing segmentation. Individual segmentation results per tile are then automatically merged via consensus analysis and then stitched to complete the segmentation for the entire image stack. SEGMENT3D is a comprehensive application that can be applied to other 3D imaging modalities and general objects. It also provides an easy way to create supervised data to advance segmentation using machine learning models.
Cell Segmentation in 3D Confocal Images using Supervoxel Merge-Forests with CNN-based Hypothesis Selection
Oct 18, 2017



Abstract:Automated segmentation approaches are crucial to quantitatively analyze large-scale 3D microscopy images. Particularly in deep tissue regions, automatic methods still fail to provide error-free segmentations. To improve the segmentation quality throughout imaged samples, we present a new supervoxel-based 3D segmentation approach that outperforms current methods and reduces the manual correction effort. The algorithm consists of gentle preprocessing and a conservative super-voxel generation method followed by supervoxel agglomeration based on local signal properties and a postprocessing step to fix under-segmentation errors using a Convolutional Neural Network. We validate the functionality of the algorithm on manually labeled 3D confocal images of the plant Arabidopis thaliana and compare the results to a state-of-the-art meristem segmentation algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge