Thiago V. Spina
CNN-based Preprocessing to Optimize Watershed-based Cell Segmentation in 3D Confocal Microscopy Images
Oct 16, 2018



Abstract:The quantitative analysis of cellular membranes helps understanding developmental processes at the cellular level. Particularly 3D microscopic image data offers valuable insights into cell dynamics, but error-free automatic segmentation remains challenging due to the huge amount of data generated and strong variations in image intensities. In this paper, we propose a new 3D segmentation approach which combines the discriminative power of convolutional neural networks (CNNs) for preprocessing and investigates the performance of three watershed-based postprocessing strategies (WS), which are well suited to segment object shapes, even when supplied with vague seed and boundary constraints. To leverage the full potential of the watershed algorithm, the multi-instance segmentation problem is initially interpreted as three-class semantic segmentation problem, which in turn is well-suited for the application of CNNs. Using manually annotated 3D confocal microscopy images of Arabidopsis thaliana, we show the superior performance of the proposed method compared to the state of the art.
SEGMENT3D: A Web-based Application for Collaborative Segmentation of 3D images used in the Shoot Apical Meristem
Oct 26, 2017



Abstract:The quantitative analysis of 3D confocal microscopy images of the shoot apical meristem helps understanding the growth process of some plants. Cell segmentation in these images is crucial for computational plant analysis and many automated methods have been proposed. However, variations in signal intensity across the image mitigate the effectiveness of those approaches with no easy way for user correction. We propose a web-based collaborative 3D image segmentation application, SEGMENT3D, to leverage automatic segmentation results. The image is divided into 3D tiles that can be either segmented interactively from scratch or corrected from a pre-existing segmentation. Individual segmentation results per tile are then automatically merged via consensus analysis and then stitched to complete the segmentation for the entire image stack. SEGMENT3D is a comprehensive application that can be applied to other 3D imaging modalities and general objects. It also provides an easy way to create supervised data to advance segmentation using machine learning models.
Cell Segmentation in 3D Confocal Images using Supervoxel Merge-Forests with CNN-based Hypothesis Selection
Oct 18, 2017



Abstract:Automated segmentation approaches are crucial to quantitatively analyze large-scale 3D microscopy images. Particularly in deep tissue regions, automatic methods still fail to provide error-free segmentations. To improve the segmentation quality throughout imaged samples, we present a new supervoxel-based 3D segmentation approach that outperforms current methods and reduces the manual correction effort. The algorithm consists of gentle preprocessing and a conservative super-voxel generation method followed by supervoxel agglomeration based on local signal properties and a postprocessing step to fix under-segmentation errors using a Convolutional Neural Network. We validate the functionality of the algorithm on manually labeled 3D confocal images of the plant Arabidopis thaliana and compare the results to a state-of-the-art meristem segmentation algorithm.
Video Human Segmentation using Fuzzy Object Models and its Application to Body Pose Estimation of Toddlers for Behavior Studies
May 29, 2013

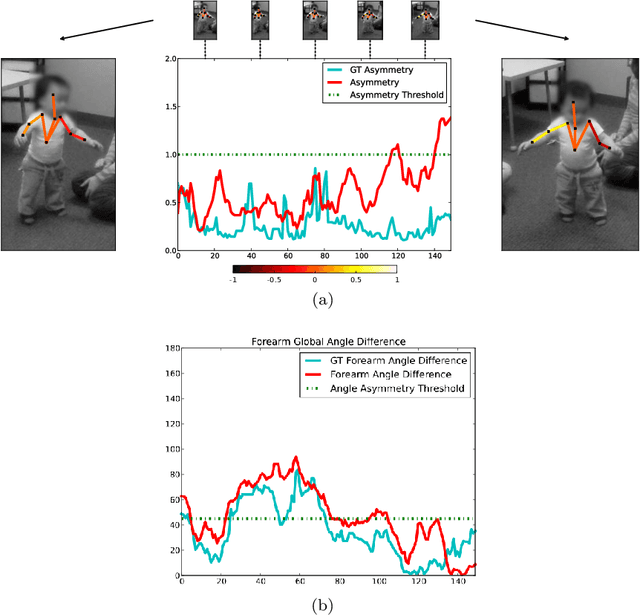

Abstract:Video object segmentation is a challenging problem due to the presence of deformable, connected, and articulated objects, intra- and inter-object occlusions, object motion, and poor lighting. Some of these challenges call for object models that can locate a desired object and separate it from its surrounding background, even when both share similar colors and textures. In this work, we extend a fuzzy object model, named cloud system model (CSM), to handle video segmentation, and evaluate it for body pose estimation of toddlers at risk of autism. CSM has been successfully used to model the parts of the brain (cerebrum, left and right brain hemispheres, and cerebellum) in order to automatically locate and separate them from each other, the connected brain stem, and the background in 3D MR-images. In our case, the objects are articulated parts (2D projections) of the human body, which can deform, cause self-occlusions, and move along the video. The proposed CSM extension handles articulation by connecting the individual clouds, body parts, of the system using a 2D stickman model. The stickman representation naturally allows us to extract 2D body pose measures of arm asymmetry patterns during unsupported gait of toddlers, a possible behavioral marker of autism. The results show that our method can provide insightful knowledge to assist the specialist's observations during real in-clinic assessments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge