Aidan Gomez
Aya Vision: Advancing the Frontier of Multilingual Multimodality
May 13, 2025Abstract:Building multimodal language models is fundamentally challenging: it requires aligning vision and language modalities, curating high-quality instruction data, and avoiding the degradation of existing text-only capabilities once vision is introduced. These difficulties are further magnified in the multilingual setting, where the need for multimodal data in different languages exacerbates existing data scarcity, machine translation often distorts meaning, and catastrophic forgetting is more pronounced. To address the aforementioned challenges, we introduce novel techniques spanning both data and modeling. First, we develop a synthetic annotation framework that curates high-quality, diverse multilingual multimodal instruction data, enabling Aya Vision models to produce natural, human-preferred responses to multimodal inputs across many languages. Complementing this, we propose a cross-modal model merging technique that mitigates catastrophic forgetting, effectively preserving text-only capabilities while simultaneously enhancing multimodal generative performance. Aya-Vision-8B achieves best-in-class performance compared to strong multimodal models such as Qwen-2.5-VL-7B, Pixtral-12B, and even much larger Llama-3.2-90B-Vision. We further scale this approach with Aya-Vision-32B, which outperforms models more than twice its size, such as Molmo-72B and LLaMA-3.2-90B-Vision. Our work advances multilingual progress on the multi-modal frontier, and provides insights into techniques that effectively bend the need for compute while delivering extremely high performance.
Command A: An Enterprise-Ready Large Language Model
Apr 01, 2025



Abstract:In this report we describe the development of Command A, a powerful large language model purpose-built to excel at real-world enterprise use cases. Command A is an agent-optimised and multilingual-capable model, with support for 23 languages of global business, and a novel hybrid architecture balancing efficiency with top of the range performance. It offers best-in-class Retrieval Augmented Generation (RAG) capabilities with grounding and tool use to automate sophisticated business processes. These abilities are achieved through a decentralised training approach, including self-refinement algorithms and model merging techniques. We also include results for Command R7B which shares capability and architectural similarities to Command A. Weights for both models have been released for research purposes. This technical report details our original training pipeline and presents an extensive evaluation of our models across a suite of enterprise-relevant tasks and public benchmarks, demonstrating excellent performance and efficiency.
Aya Expanse: Combining Research Breakthroughs for a New Multilingual Frontier
Dec 05, 2024



Abstract:We introduce the Aya Expanse model family, a new generation of 8B and 32B parameter multilingual language models, aiming to address the critical challenge of developing highly performant multilingual models that match or surpass the capabilities of monolingual models. By leveraging several years of research at Cohere For AI and Cohere, including advancements in data arbitrage, multilingual preference training, and model merging, Aya Expanse sets a new state-of-the-art in multilingual performance. Our evaluations on the Arena-Hard-Auto dataset, translated into 23 languages, demonstrate that Aya Expanse 8B and 32B outperform leading open-weight models in their respective parameter classes, including Gemma 2, Qwen 2.5, and Llama 3.1, achieving up to a 76.6% win-rate. Notably, Aya Expanse 32B outperforms Llama 3.1 70B, a model with twice as many parameters, achieving a 54.0% win-rate. In this short technical report, we present extended evaluation results for the Aya Expanse model family and release their open-weights, together with a new multilingual evaluation dataset m-ArenaHard.
Tranception: protein fitness prediction with autoregressive transformers and inference-time retrieval
May 27, 2022



Abstract:The ability to accurately model the fitness landscape of protein sequences is critical to a wide range of applications, from quantifying the effects of human variants on disease likelihood, to predicting immune-escape mutations in viruses and designing novel biotherapeutic proteins. Deep generative models of protein sequences trained on multiple sequence alignments have been the most successful approaches so far to address these tasks. The performance of these methods is however contingent on the availability of sufficiently deep and diverse alignments for reliable training. Their potential scope is thus limited by the fact many protein families are hard, if not impossible, to align. Large language models trained on massive quantities of non-aligned protein sequences from diverse families address these problems and show potential to eventually bridge the performance gap. We introduce Tranception, a novel transformer architecture leveraging autoregressive predictions and retrieval of homologous sequences at inference to achieve state-of-the-art fitness prediction performance. Given its markedly higher performance on multiple mutants, robustness to shallow alignments and ability to score indels, our approach offers significant gain of scope over existing approaches. To enable more rigorous model testing across a broader range of protein families, we develop ProteinGym -- an extensive set of multiplexed assays of variant effects, substantially increasing both the number and diversity of assays compared to existing benchmarks.
The Difficulty of Training Sparse Neural Networks
Jul 17, 2019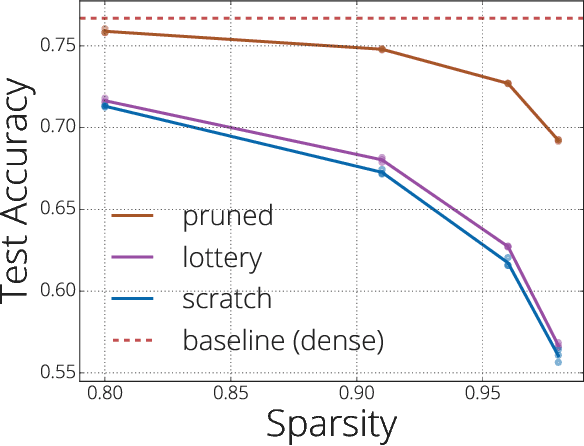
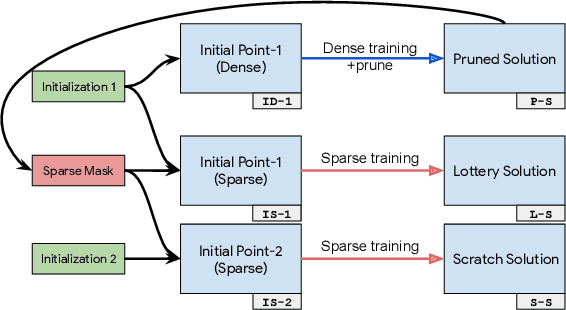
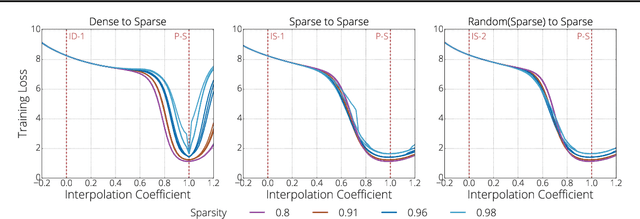
Abstract:We investigate the difficulties of training sparse neural networks and make new observations about optimization dynamics and the energy landscape within the sparse regime. Recent work of \citep{Gale2019, Liu2018} has shown that sparse ResNet-50 architectures trained on ImageNet-2012 dataset converge to solutions that are significantly worse than those found by pruning. We show that, despite the failure of optimizers, there is a linear path with a monotonically decreasing objective from the initialization to the "good" solution. Additionally, our attempts to find a decreasing objective path from "bad" solutions to the "good" ones in the sparse subspace fail. However, if we allow the path to traverse the dense subspace, then we consistently find a path between two solutions. These findings suggest traversing extra dimensions may be needed to escape stationary points found in the sparse subspace.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge