Zhiyi Kuang
Learning Game-Playing Agents with Generative Code Optimization
Aug 27, 2025Abstract:We present a generative optimization approach for learning game-playing agents, where policies are represented as Python programs and refined using large language models (LLMs). Our method treats decision-making policies as self-evolving code, with current observation as input and an in-game action as output, enabling agents to self-improve through execution traces and natural language feedback with minimal human intervention. Applied to Atari games, our game-playing Python program achieves performance competitive with deep reinforcement learning (RL) baselines while using significantly less training time and much fewer environment interactions. This work highlights the promise of programmatic policy representations for building efficient, adaptable agents capable of complex, long-horizon reasoning.
MonoHair: High-Fidelity Hair Modeling from a Monocular Video
Mar 27, 2024



Abstract:Undoubtedly, high-fidelity 3D hair is crucial for achieving realism, artistic expression, and immersion in computer graphics. While existing 3D hair modeling methods have achieved impressive performance, the challenge of achieving high-quality hair reconstruction persists: they either require strict capture conditions, making practical applications difficult, or heavily rely on learned prior data, obscuring fine-grained details in images. To address these challenges, we propose MonoHair,a generic framework to achieve high-fidelity hair reconstruction from a monocular video, without specific requirements for environments. Our approach bifurcates the hair modeling process into two main stages: precise exterior reconstruction and interior structure inference. The exterior is meticulously crafted using our Patch-based Multi-View Optimization (PMVO). This method strategically collects and integrates hair information from multiple views, independent of prior data, to produce a high-fidelity exterior 3D line map. This map not only captures intricate details but also facilitates the inference of the hair's inner structure. For the interior, we employ a data-driven, multi-view 3D hair reconstruction method. This method utilizes 2D structural renderings derived from the reconstructed exterior, mirroring the synthetic 2D inputs used during training. This alignment effectively bridges the domain gap between our training data and real-world data, thereby enhancing the accuracy and reliability of our interior structure inference. Lastly, we generate a strand model and resolve the directional ambiguity by our hair growth algorithm. Our experiments demonstrate that our method exhibits robustness across diverse hairstyles and achieves state-of-the-art performance. For more results, please refer to our project page https://keyuwu-cs.github.io/MonoHair/.
Multimodality Helps Unimodality: Cross-Modal Few-Shot Learning with Multimodal Models
Jan 18, 2023



Abstract:The ability to quickly learn a new task with minimal instruction - known as few-shot learning - is a central aspect of intelligent agents. Classical few-shot benchmarks make use of few-shot samples from a single modality, but such samples may not be sufficient to characterize an entire concept class. In contrast, humans use cross-modal information to learn new concepts efficiently. In this work, we demonstrate that one can indeed build a better ${\bf visual}$ dog classifier by ${\bf read}$ing about dogs and ${\bf listen}$ing to them bark. To do so, we exploit the fact that recent multimodal foundation models such as CLIP are inherently cross-modal, mapping different modalities to the same representation space. Specifically, we propose a simple cross-modal adaptation approach that learns from few-shot examples spanning different modalities. By repurposing class names as additional one-shot training samples, we achieve SOTA results with an embarrassingly simple linear classifier for vision-language adaptation. Furthermore, we show that our approach can benefit existing methods such as prefix tuning, adapters, and classifier ensembling. Finally, to explore other modalities beyond vision and language, we construct the first (to our knowledge) audiovisual few-shot benchmark and use cross-modal training to improve the performance of both image and audio classification.
Using Deep Learning Sequence Models to Identify SARS-CoV-2 Divergence
Nov 12, 2021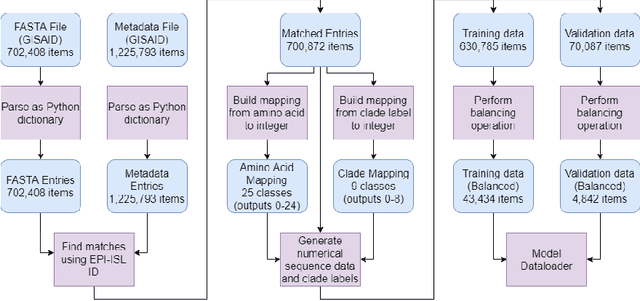
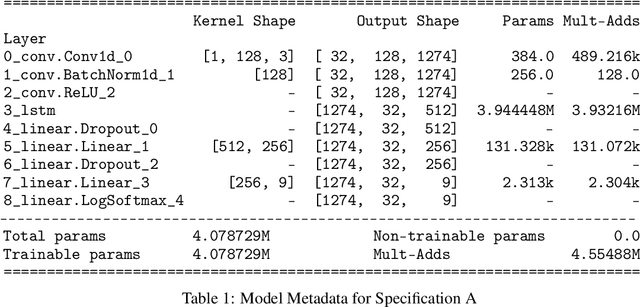
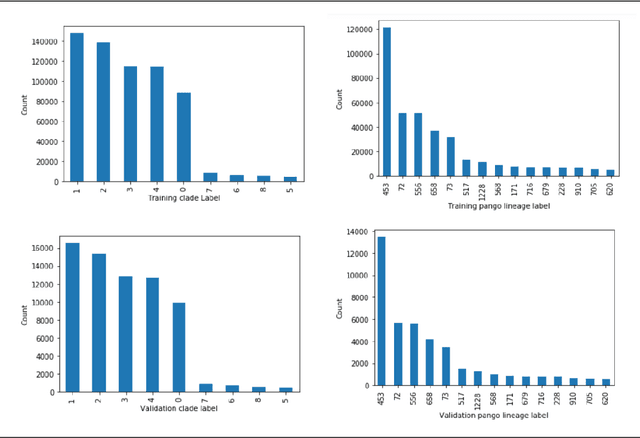
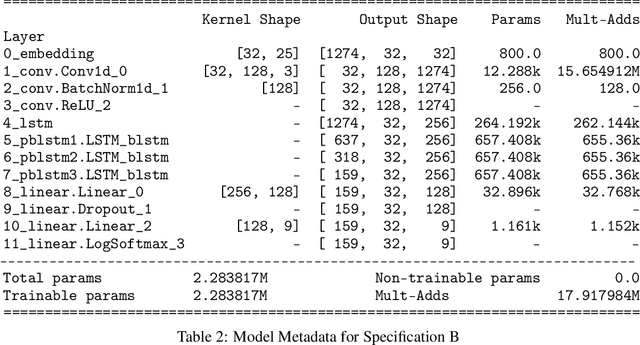
Abstract:SARS-CoV-2 is an upper respiratory system RNA virus that has caused over 3 million deaths and infecting over 150 million worldwide as of May 2021. With thousands of strains sequenced to date, SARS-CoV-2 mutations pose significant challenges to scientists on keeping pace with vaccine development and public health measures. Therefore, an efficient method of identifying the divergence of lab samples from patients would greatly aid the documentation of SARS-CoV-2 genomics. In this study, we propose a neural network model that leverages recurrent and convolutional units to directly take in amino acid sequences of spike proteins and classify corresponding clades. We also compared our model's performance with Bidirectional Encoder Representations from Transformers (BERT) pre-trained on protein database. Our approach has the potential of providing a more computationally efficient alternative to current homology based intra-species differentiation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge