Yuxin Pei
SegMix: A Simple Structure-Aware Data Augmentation Method
Nov 16, 2023



Abstract:Interpolation-based Data Augmentation (DA) methods (Mixup) linearly interpolate the inputs and labels of two or more training examples. Mixup has more recently been adapted to the field of Natural Language Processing (NLP), mainly for sequence labeling tasks. However, such a simple adoption yields mixed or unstable improvements over the baseline models. We argue that the direct-adoption methods do not account for structures in NLP tasks. To this end, we propose SegMix, a collection of interpolation-based DA algorithms that can adapt to task-specific structures. SegMix poses fewer constraints on data structures, is robust to various hyperparameter settings, applies to more task settings, and adds little computational overhead. In the algorithm's core, we apply interpolation methods on task-specific meaningful segments, in contrast to applying them on sequences as in prior work. We find SegMix to be a flexible framework that combines rule-based DA methods with interpolation-based methods, creating interesting mixtures of DA techniques. We show that SegMix consistently improves performance over strong baseline models in Named Entity Recognition (NER) and Relation Extraction (RE) tasks, especially under data-scarce settings. Furthermore, this method is easy to implement and adds negligible training overhead.
Using Deep Learning Sequence Models to Identify SARS-CoV-2 Divergence
Nov 12, 2021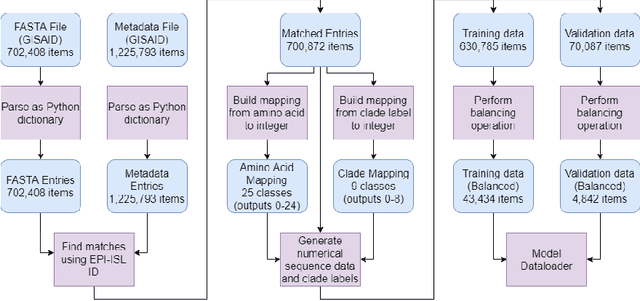
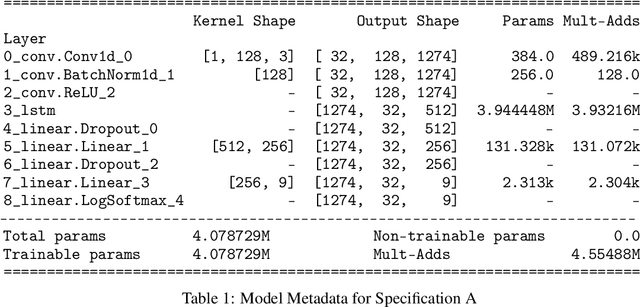
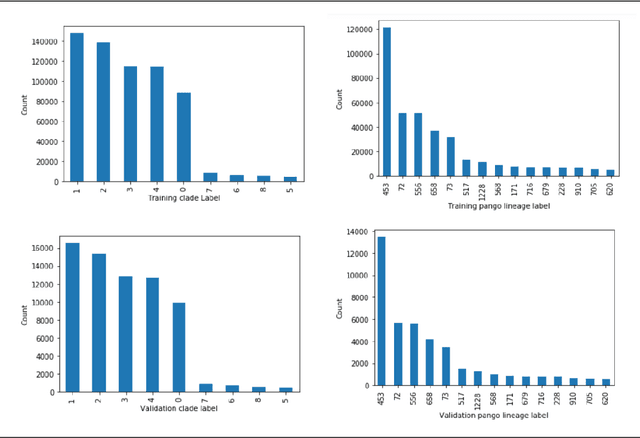
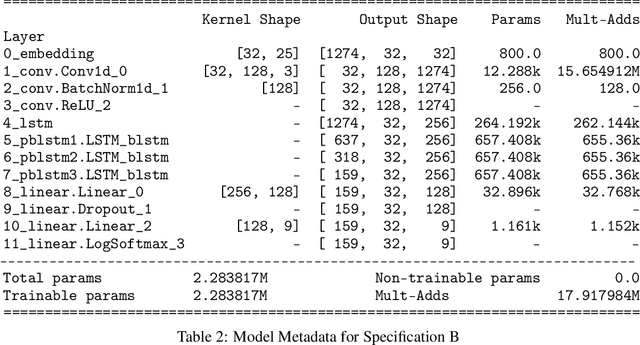
Abstract:SARS-CoV-2 is an upper respiratory system RNA virus that has caused over 3 million deaths and infecting over 150 million worldwide as of May 2021. With thousands of strains sequenced to date, SARS-CoV-2 mutations pose significant challenges to scientists on keeping pace with vaccine development and public health measures. Therefore, an efficient method of identifying the divergence of lab samples from patients would greatly aid the documentation of SARS-CoV-2 genomics. In this study, we propose a neural network model that leverages recurrent and convolutional units to directly take in amino acid sequences of spike proteins and classify corresponding clades. We also compared our model's performance with Bidirectional Encoder Representations from Transformers (BERT) pre-trained on protein database. Our approach has the potential of providing a more computationally efficient alternative to current homology based intra-species differentiation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge