Jeff Tan
CRISP: Contact-Guided Real2Sim from Monocular Video with Planar Scene Primitives
Dec 21, 2025Abstract:We introduce CRISP, a method that recovers simulatable human motion and scene geometry from monocular video. Prior work on joint human-scene reconstruction relies on data-driven priors and joint optimization with no physics in the loop, or recovers noisy geometry with artifacts that cause motion tracking policies with scene interactions to fail. In contrast, our key insight is to recover convex, clean, and simulation-ready geometry by fitting planar primitives to a point cloud reconstruction of the scene, via a simple clustering pipeline over depth, normals, and flow. To reconstruct scene geometry that might be occluded during interactions, we make use of human-scene contact modeling (e.g., we use human posture to reconstruct the occluded seat of a chair). Finally, we ensure that human and scene reconstructions are physically-plausible by using them to drive a humanoid controller via reinforcement learning. Our approach reduces motion tracking failure rates from 55.2\% to 6.9\% on human-centric video benchmarks (EMDB, PROX), while delivering a 43\% faster RL simulation throughput. We further validate it on in-the-wild videos including casually-captured videos, Internet videos, and even Sora-generated videos. This demonstrates CRISP's ability to generate physically-valid human motion and interaction environments at scale, greatly advancing real-to-sim applications for robotics and AR/VR.
MonoFusion: Sparse-View 4D Reconstruction via Monocular Fusion
Jul 31, 2025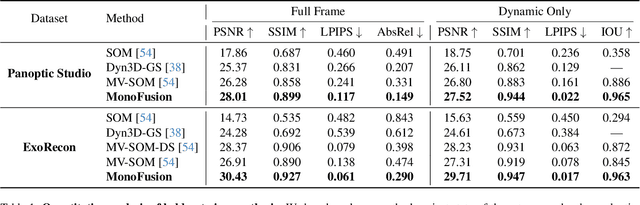
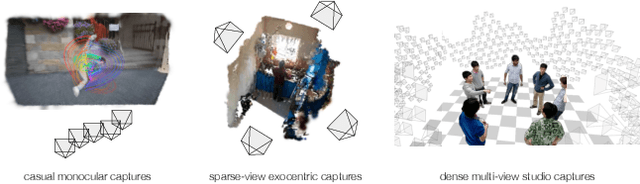
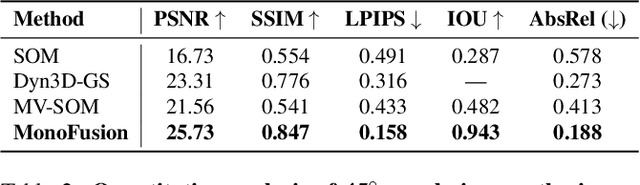
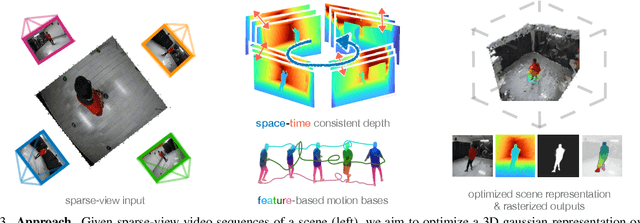
Abstract:We address the problem of dynamic scene reconstruction from sparse-view videos. Prior work often requires dense multi-view captures with hundreds of calibrated cameras (e.g. Panoptic Studio). Such multi-view setups are prohibitively expensive to build and cannot capture diverse scenes in-the-wild. In contrast, we aim to reconstruct dynamic human behaviors, such as repairing a bike or dancing, from a small set of sparse-view cameras with complete scene coverage (e.g. four equidistant inward-facing static cameras). We find that dense multi-view reconstruction methods struggle to adapt to this sparse-view setup due to limited overlap between viewpoints. To address these limitations, we carefully align independent monocular reconstructions of each camera to produce time- and view-consistent dynamic scene reconstructions. Extensive experiments on PanopticStudio and Ego-Exo4D demonstrate that our method achieves higher quality reconstructions than prior art, particularly when rendering novel views. Code, data, and data-processing scripts are available on https://github.com/ImNotPrepared/MonoFusion.
DiffusionSfM: Predicting Structure and Motion via Ray Origin and Endpoint Diffusion
May 08, 2025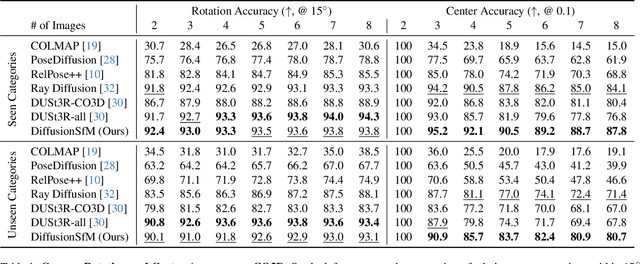
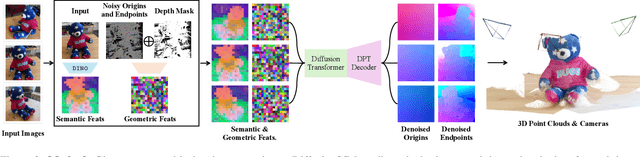

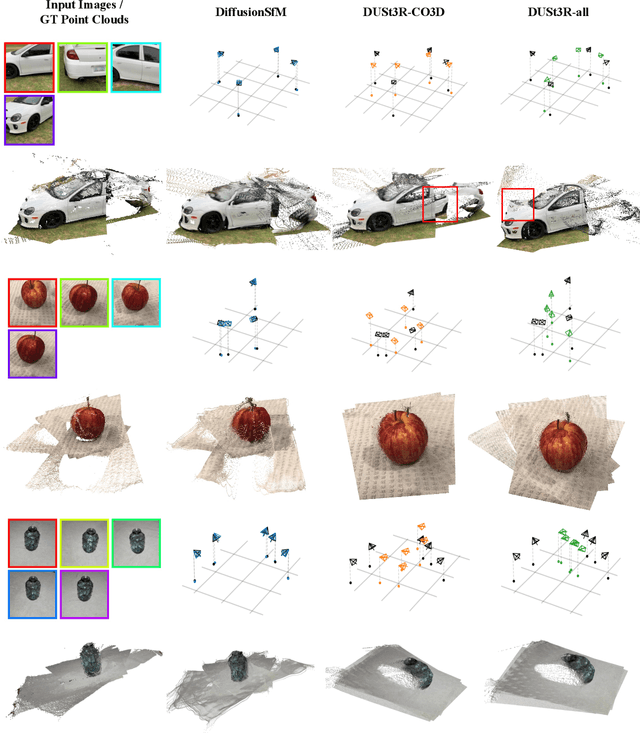
Abstract:Current Structure-from-Motion (SfM) methods typically follow a two-stage pipeline, combining learned or geometric pairwise reasoning with a subsequent global optimization step. In contrast, we propose a data-driven multi-view reasoning approach that directly infers 3D scene geometry and camera poses from multi-view images. Our framework, DiffusionSfM, parameterizes scene geometry and cameras as pixel-wise ray origins and endpoints in a global frame and employs a transformer-based denoising diffusion model to predict them from multi-view inputs. To address practical challenges in training diffusion models with missing data and unbounded scene coordinates, we introduce specialized mechanisms that ensure robust learning. We empirically validate DiffusionSfM on both synthetic and real datasets, demonstrating that it outperforms classical and learning-based approaches while naturally modeling uncertainty.
DressRecon: Freeform 4D Human Reconstruction from Monocular Video
Sep 30, 2024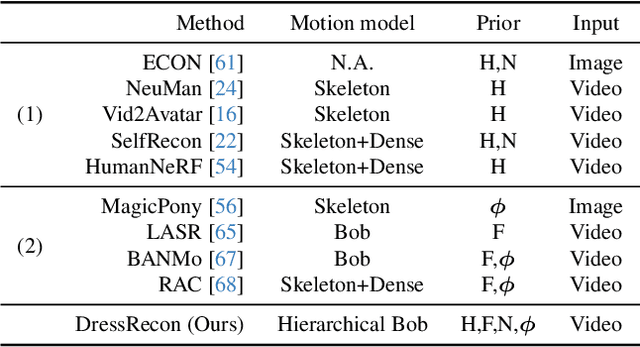
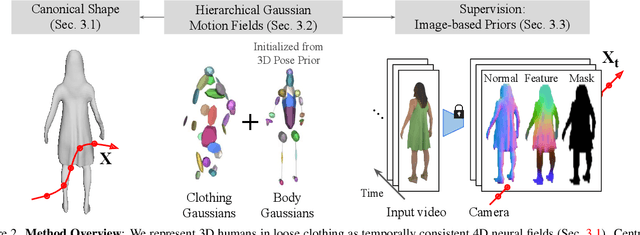
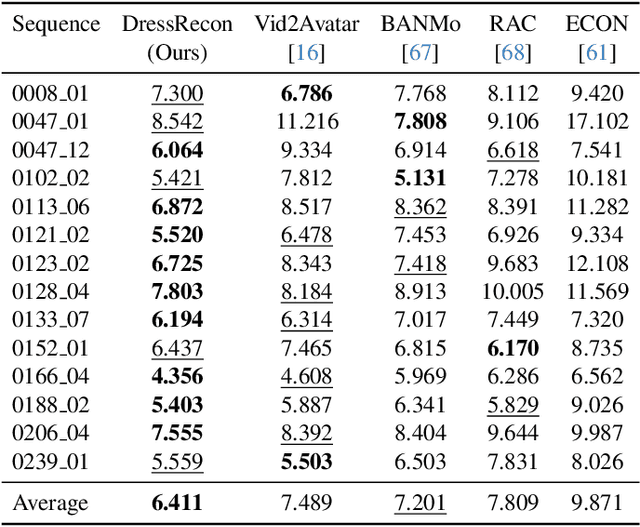
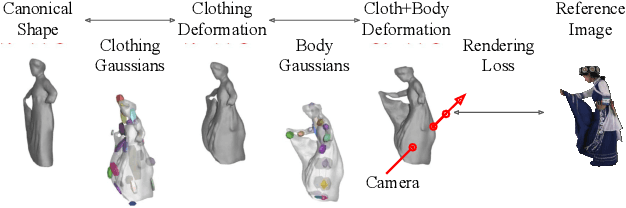
Abstract:We present a method to reconstruct time-consistent human body models from monocular videos, focusing on extremely loose clothing or handheld object interactions. Prior work in human reconstruction is either limited to tight clothing with no object interactions, or requires calibrated multi-view captures or personalized template scans which are costly to collect at scale. Our key insight for high-quality yet flexible reconstruction is the careful combination of generic human priors about articulated body shape (learned from large-scale training data) with video-specific articulated "bag-of-bones" deformation (fit to a single video via test-time optimization). We accomplish this by learning a neural implicit model that disentangles body versus clothing deformations as separate motion model layers. To capture subtle geometry of clothing, we leverage image-based priors such as human body pose, surface normals, and optical flow during optimization. The resulting neural fields can be extracted into time-consistent meshes, or further optimized as explicit 3D Gaussians for high-fidelity interactive rendering. On datasets with highly challenging clothing deformations and object interactions, DressRecon yields higher-fidelity 3D reconstructions than prior art. Project page: https://jefftan969.github.io/dressrecon/
Using Deep Learning Sequence Models to Identify SARS-CoV-2 Divergence
Nov 12, 2021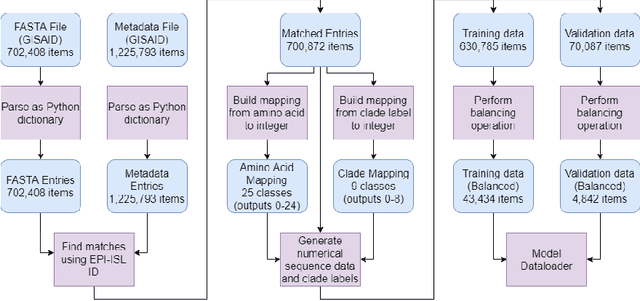
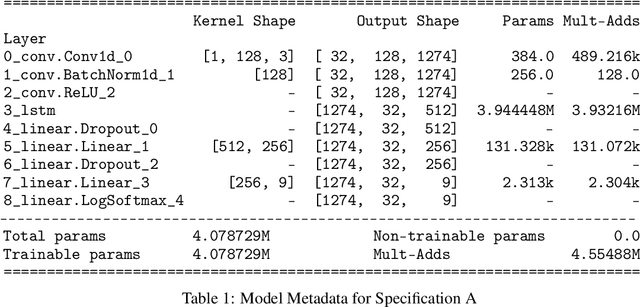
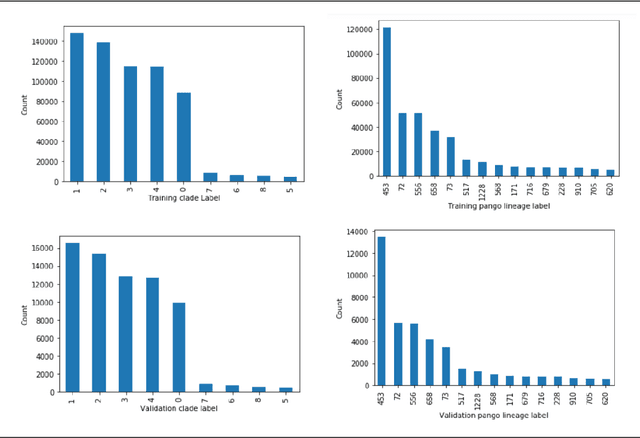
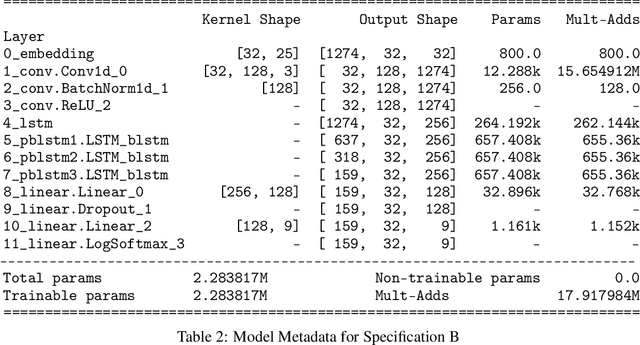
Abstract:SARS-CoV-2 is an upper respiratory system RNA virus that has caused over 3 million deaths and infecting over 150 million worldwide as of May 2021. With thousands of strains sequenced to date, SARS-CoV-2 mutations pose significant challenges to scientists on keeping pace with vaccine development and public health measures. Therefore, an efficient method of identifying the divergence of lab samples from patients would greatly aid the documentation of SARS-CoV-2 genomics. In this study, we propose a neural network model that leverages recurrent and convolutional units to directly take in amino acid sequences of spike proteins and classify corresponding clades. We also compared our model's performance with Bidirectional Encoder Representations from Transformers (BERT) pre-trained on protein database. Our approach has the potential of providing a more computationally efficient alternative to current homology based intra-species differentiation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge