Zhijie Wen
Open set label noise learning with robust sample selection and margin-guided module
Jan 08, 2025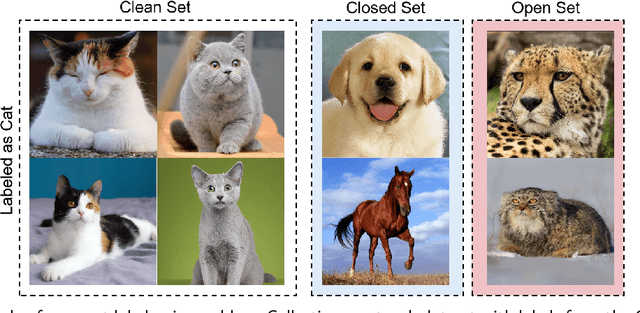
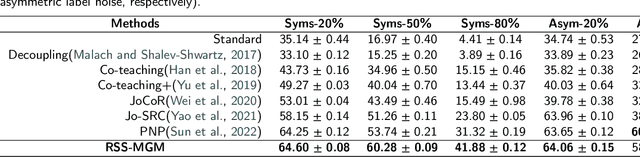
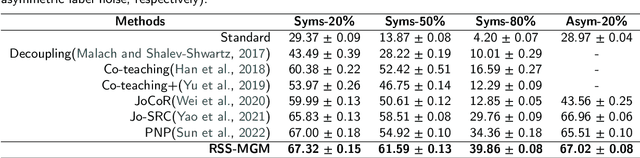
Abstract:In recent years, the remarkable success of deep neural networks (DNNs) in computer vision is largely due to large-scale, high-quality labeled datasets. Training directly on real-world datasets with label noise may result in overfitting. The traditional method is limited to deal with closed set label noise, where noisy training data has true class labels within the known label space. However, there are some real-world datasets containing open set label noise, which means that some samples belong to an unknown class outside the known label space. To address the open set label noise problem, we introduce a method based on Robust Sample Selection and Margin-Guided Module (RSS-MGM). Firstly, unlike the prior clean sample selection approach, which only select a limited number of clean samples, a robust sample selection module combines small loss selection or high-confidence sample selection to obtain more clean samples. Secondly, to efficiently distinguish open set label noise and closed set ones, margin functions are designed to filter open-set data and closed set data. Thirdly, different processing methods are selected for different types of samples in order to fully utilize the data's prior information and optimize the whole model. Furthermore, extensive experimental results with noisy labeled data from benchmark datasets and real-world datasets, such as CIFAR-100N-C, CIFAR80N-O, WebFG-469, and Food101N, indicate that our approach outperforms many state-of-the-art label noise learning methods. Especially, it can more accurately divide open set label noise samples and closed set ones.
Re-Visible Dual-Domain Self-Supervised Deep Unfolding Network for MRI Reconstruction
Jan 07, 2025



Abstract:Magnetic Resonance Imaging (MRI) is widely used in clinical practice, but suffered from prolonged acquisition time. Although deep learning methods have been proposed to accelerate acquisition and demonstrate promising performance, they rely on high-quality fully-sampled datasets for training in a supervised manner. However, such datasets are time-consuming and expensive-to-collect, which constrains their broader applications. On the other hand, self-supervised methods offer an alternative by enabling learning from under-sampled data alone, but most existing methods rely on further partitioned under-sampled k-space data as model's input for training, resulting in a loss of valuable information. Additionally, their models have not fully incorporated image priors, leading to degraded reconstruction performance. In this paper, we propose a novel re-visible dual-domain self-supervised deep unfolding network to address these issues when only under-sampled datasets are available. Specifically, by incorporating re-visible dual-domain loss, all under-sampled k-space data are utilized during training to mitigate information loss caused by further partitioning. This design enables the model to implicitly adapt to all under-sampled k-space data as input. Additionally, we design a deep unfolding network based on Chambolle and Pock Proximal Point Algorithm (DUN-CP-PPA) to achieve end-to-end reconstruction, incorporating imaging physics and image priors to guide the reconstruction process. By employing a Spatial-Frequency Feature Extraction (SFFE) block to capture global and local feature representation, we enhance the model's efficiency to learn comprehensive image priors. Experiments conducted on the fastMRI and IXI datasets demonstrate that our method significantly outperforms state-of-the-art approaches in terms of reconstruction performance.
Deep Unfolding Network with Spatial Alignment for multi-modal MRI reconstruction
Dec 28, 2023



Abstract:Multi-modal Magnetic Resonance Imaging (MRI) offers complementary diagnostic information, but some modalities are limited by the long scanning time. To accelerate the whole acquisition process, MRI reconstruction of one modality from highly undersampled k-space data with another fully-sampled reference modality is an efficient solution. However, the misalignment between modalities, which is common in clinic practice, can negatively affect reconstruction quality. Existing deep learning-based methods that account for inter-modality misalignment perform better, but still share two main common limitations: (1) The spatial alignment task is not adaptively integrated with the reconstruction process, resulting in insufficient complementarity between the two tasks; (2) the entire framework has weak interpretability. In this paper, we construct a novel Deep Unfolding Network with Spatial Alignment, termed DUN-SA, to appropriately embed the spatial alignment task into the reconstruction process. Concretely, we derive a novel joint alignment-reconstruction model with a specially designed cross-modal spatial alignment term. By relaxing the model into cross-modal spatial alignment and multi-modal reconstruction tasks, we propose an effective algorithm to solve this model alternatively. Then, we unfold the iterative steps of the proposed algorithm and design corresponding network modules to build DUN-SA with interpretability. Through end-to-end training, we effectively compensate for spatial misalignment using only reconstruction loss, and utilize the progressively aligned reference modality to provide inter-modality prior to improve the reconstruction of the target modality. Comprehensive experiments on three real datasets demonstrate that our method exhibits superior reconstruction performance compared to state-of-the-art methods.
Spatial and Modal Optimal Transport for Fast Cross-Modal MRI Reconstruction
May 04, 2023



Abstract:Multi-modal Magnetic Resonance Imaging (MRI) plays an important role in clinical medicine. However, the acquisitions of some modalities, such as the T2-weighted modality, need a long time and they are always accompanied by motion artifacts. On the other hand, the T1-weighted image (T1WI) shares the same underlying information with T2-weighted image (T2WI), which needs a shorter scanning time. Therefore, in this paper we accelerate the acquisition of the T2WI by introducing the auxiliary modality (T1WI). Concretely, we first reconstruct high-quality T2WIs with under-sampled T2WIs. Here, we realize fast T2WI reconstruction by reducing the sampling rate in the k-space. Second, we establish a cross-modal synthesis task to generate the synthetic T2WIs for guiding better T2WI reconstruction. Here, we obtain the synthetic T2WIs by decomposing the whole cross-modal generation mapping into two OT processes, the spatial alignment mapping on the T1 image manifold and the cross-modal synthesis mapping from aligned T1WIs to T2WIs. It overcomes the negative transfer caused by the spatial misalignment. Then, we prove the reconstruction and the synthesis tasks are well complementary. Finally, we compare it with state-of-the-art approaches on an open dataset FastMRI and an in-house dataset to testify the validity of the proposed method.
Learning Discriminative Representations for Fine-Grained Diabetic Retinopathy Grading
Nov 04, 2020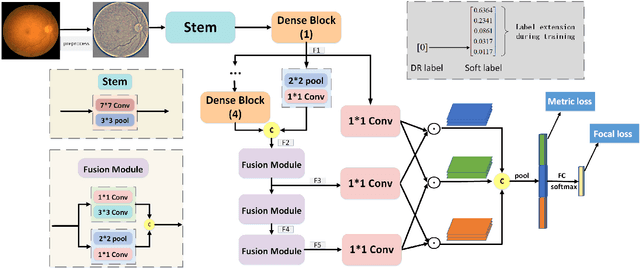
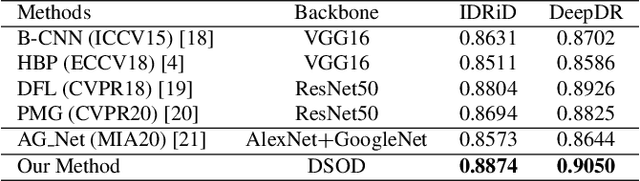
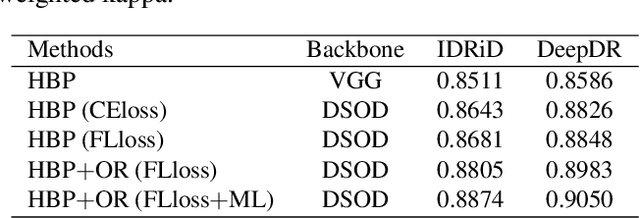
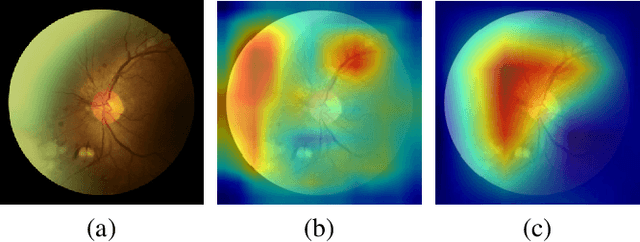
Abstract:Diabetic retinopathy (DR) is one of the leading causes of blindness. However, no specific symptoms of early DR lead to a delayed diagnosis, which results in disease progression in patients. To determine the disease severity levels, ophthalmologists need to focus on the discriminative parts of the fundus images. In recent years, deep learning has achieved great success in medical image analysis. However, most works directly employ algorithms based on convolutional neural networks (CNNs), which ignore the fact that the difference among classes is subtle and gradual. Hence, we consider automatic image grading of DR as a fine-grained classification task, and construct a bilinear model to identify the pathologically discriminative areas. In order to leverage the ordinal information among classes, we use an ordinal regression method to obtain the soft labels. In addition, other than only using a categorical loss to train our network, we also introduce the metric loss to learn a more discriminative feature space. Experimental results demonstrate the superior performance of the proposed method on two public IDRiD and DeepDR datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge