Zhenzhang Li
MpoxMamba: A Grouped Mamba-based Lightweight Hybrid Network for Mpox Detection
Sep 06, 2024



Abstract:Due to the lack of effective mpox detection tools, the mpox virus continues to spread worldwide and has once again been declared a public health emergency of international concern by the World Health Organization. Deep learning-based mpox detection tools are crucial to alleviate mpox outbreak. However, existing methods have difficulty in achieving a good trade-off between detection performance, parameter size, and model complexity, which is crucial for practical applications and widespread deployment, especially in resource-limited scenarios. Given that the success of Mamba in modeling long-range dependencies and its linear complexity, we proposed a lightweight hybrid architecture called MpoxMamba. MpoxMamba utilizes deep separable convolutions to extract local feature representations in mpox skin lesions, and greatly enhances the model's ability to model the global contextual information by grouped Mamba modules. Experimental results on two widely recognized mpox datasets demonstrate that MpoxMamba outperforms existing mpox detection methods and state-of-the-art lightweight models. We also developed a web-based online application to provide free mpox detection services to the public in the epidemic areas (http://5227i971s5.goho.co:30290). The source codes of MpoxMamba are available at https://github.com/YubiaoYue/MpoxMamba.
Out-of-distribution Detection in Medical Image Analysis: A survey
Apr 28, 2024
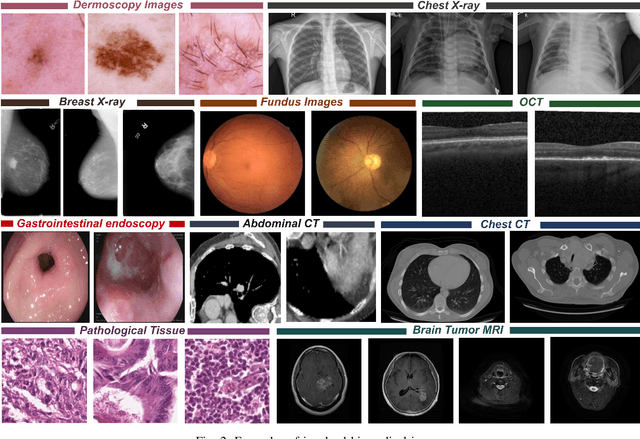
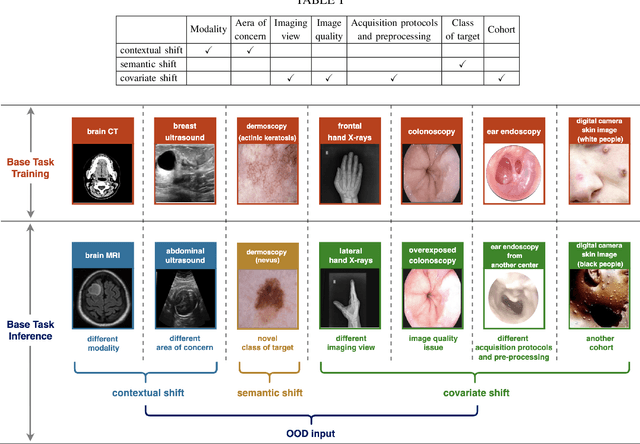

Abstract:Computer-aided diagnostics has benefited from the development of deep learning-based computer vision techniques in these years. Traditional supervised deep learning methods assume that the test sample is drawn from the identical distribution as the training data. However, it is possible to encounter out-of-distribution samples in real-world clinical scenarios, which may cause silent failure in deep learning-based medical image analysis tasks. Recently, research has explored various out-of-distribution (OOD) detection situations and techniques to enable a trustworthy medical AI system. In this survey, we systematically review the recent advances in OOD detection in medical image analysis. We first explore several factors that may cause a distributional shift when using a deep-learning-based model in clinic scenarios, with three different types of distributional shift well defined on top of these factors. Then a framework is suggested to categorize and feature existing solutions, while the previous studies are reviewed based on the methodology taxonomy. Our discussion also includes evaluation protocols and metrics, as well as the challenge and a research direction lack of exploration.
MedMamba: Vision Mamba for Medical Image Classification
Mar 06, 2024

Abstract:Medical image classification is a very fundamental and crucial task in the field of computer vision. These years, CNN-based and Transformer-based models are widely used in classifying various medical images. Unfortunately, The limitation of CNNs in long-range modeling capabilities prevent them from effectively extracting fine-grained features in medical images , while Transformers are hampered by their quadratic computational complexity. Recent research has shown that the state space model (SSM) represented by Mamba can efficiently model long-range interactions while maintaining linear computational complexity. Inspired by this, we propose Vision Mamba for medical image classification (MedMamba). More specifically, we introduce a novel Conv-SSM module, which combines the local feature extraction ability of convolutional layers with the ability of SSM to capture long-range dependency. To demonstrate the potential of MedMamba, we conduct extensive experiments using three publicly available medical datasets with different imaging techniques (i.e., Kvasir (endoscopic images), FETAL_PLANES_DB (ultrasound images) and Covid19-Pneumonia-Normal Chest X-Ray (X-ray images)) and two private datasets built by ourselves. Experimental results show that the proposed MedMamba performs well in detecting lesions in various medical images. To the best of our knowledge, this is the first Vision Mamba tailored for medical image classification. The purpose of this work is to establish a new baseline for medical image classification tasks and provide valuable insights for the future development of more efficient and effective SSM-based artificial intelligence algorithms and application systems in the medical. Source code has been available at https://github.com/YubiaoYue/MedMamba.
Adversarial Masked Image Inpainting for Robust Detection of Mpox and Non-Mpox
Oct 10, 2023Abstract:Due to the lack of efficient mpox diagnostic technology, mpox cases continue to increase. Recently, the great potential of deep learning models in detecting mpox and non-mpox has been proven. However, existing models learn image representations via image classification, which results in they may be easily susceptible to interference from real-world noise, require diverse non-mpox images, and fail to detect abnormal input. These drawbacks make classification models inapplicable in real-world settings. To address these challenges, we propose "Mask, Inpainting, and Measure" (MIM). In MIM's pipeline, a generative adversarial network only learns mpox image representations by inpainting the masked mpox images. Then, MIM determines whether the input belongs to mpox by measuring the similarity between the inpainted image and the original image. The underlying intuition is that since MIM solely models mpox images, it struggles to accurately inpaint non-mpox images in real-world settings. Without utilizing any non-mpox images, MIM cleverly detects mpox and non-mpox and can handle abnormal inputs. We used the recognized mpox dataset (MSLD) and images of eighteen non-mpox skin diseases to verify the effectiveness and robustness of MIM. Experimental results show that the average AUROC of MIM achieves 0.8237. In addition, we demonstrated the drawbacks of classification models and buttressed the potential of MIM through clinical validation. Finally, we developed an online smartphone app to provide free testing to the public in affected areas. This work first employs generative models to improve mpox detection and provides new insights into binary decision-making tasks in medical images.
U-SEANNet: A Simple, Efficient and Applied U-Shaped Network for Diagnosis of Nasal Diseases on Nasal Endoscopic Images
Sep 02, 2023Abstract:Numerous studies have affirmed that deep learning models can facilitate early diagnosis of lesions in endoscopic images. However, the lack of available datasets stymies advancements in research on nasal endoscopy, and existing models fail to strike a good trade-off between model diagnosis performance, model complexity and parameters size, rendering them unsuitable for real-world application. To bridge these gaps, we created the first large-scale nasal endoscopy dataset, named 7-NasalEID, comprising 11,352 images that contain six common nasal diseases and normal samples. Subsequently, we proposed U-SEANNet, an innovative U-shaped architecture, underpinned by depth-wise separable convolution. Moreover, to enhance its capacity for detecting nuanced discrepancies in input images, U-SEANNet employs the Global-Local Channel Feature Fusion module, enabling it to utilize salient channel features from both global and local contexts. To demonstrate U-SEANNet's potential, we benchmarked U-SEANNet against seventeen modern architectures through five-fold cross-validation. The experimental results show that U-SEANNet achieves a commendable accuracy of 93.58%. Notably, U-SEANNet's parameters size and GFLOPs are only 0.78M and 0.21, respectively. Our findings suggest U-SEANNet is the state-of-the-art model for nasal diseases diagnosis in endoscopic images.
US-SFNet: A Spatial-Frequency Domain-based Multi-branch Network for Cervical Lymph Node Lesions Diagnoses in Ultrasound Images
Aug 31, 2023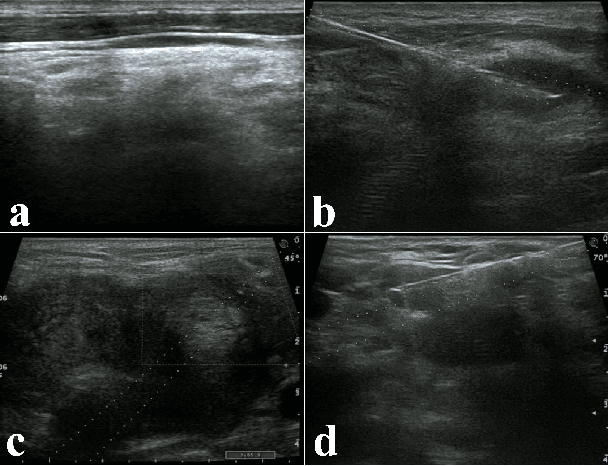
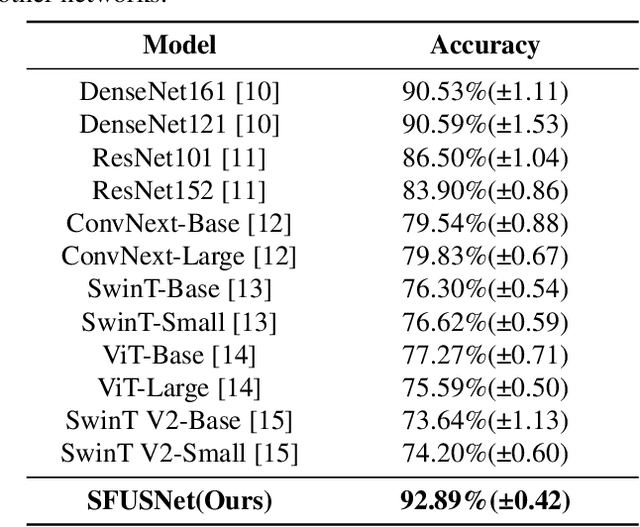
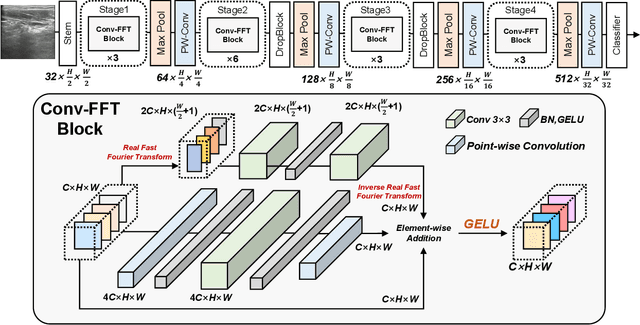
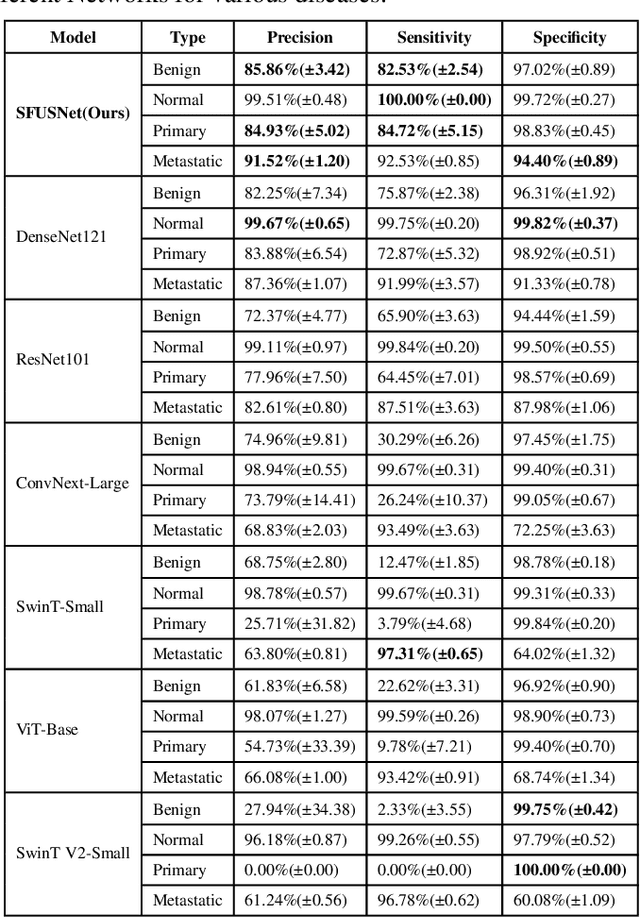
Abstract:Ultrasound imaging serves as a pivotal tool for diagnosing cervical lymph node lesions. However, the diagnoses of these images largely hinge on the expertise of medical practitioners, rendering the process susceptible to misdiagnoses. Although rapidly developing deep learning has substantially improved the diagnoses of diverse ultrasound images, there remains a conspicuous research gap concerning cervical lymph nodes. The objective of our work is to accurately diagnose cervical lymph node lesions by leveraging a deep learning model. To this end, we first collected 3392 images containing normal lymph nodes, benign lymph node lesions, malignant primary lymph node lesions, and malignant metastatic lymph node lesions. Given that ultrasound images are generated by the reflection and scattering of sound waves across varied bodily tissues, we proposed the Conv-FFT Block. It integrates convolutional operations with the fast Fourier transform to more astutely model the images. Building upon this foundation, we designed a novel architecture, named US-SFNet. This architecture not only discerns variances in ultrasound images from the spatial domain but also adeptly captures microstructural alterations across various lesions in the frequency domain. To ascertain the potential of US-SFNet, we benchmarked it against 12 popular architectures through five-fold cross-validation. The results show that US-SFNet is SOTA and can achieve 92.89% accuracy, 90.46% precision, 89.95% sensitivity and 97.49% specificity, respectively.
Ultrafast-and-Ultralight ConvNet-Based Intelligent Monitoring System for Diagnosing Early-Stage Mpox Anytime and Anywhere
Aug 25, 2023Abstract:Due to the lack of more efficient diagnostic tools for monkeypox, its spread remains unchecked, presenting a formidable challenge to global health. While the high efficacy of deep learning models for monkeypox diagnosis has been demonstrated in related studies, the overlook of inference speed, the parameter size and diagnosis performance for early-stage monkeypox renders the models inapplicable in real-world settings. To address these challenges, we proposed an ultrafast and ultralight network named Fast-MpoxNet. Fast-MpoxNet possesses only 0.27M parameters and can process input images at 68 frames per second (FPS) on the CPU. To counteract the diagnostic performance limitation brought about by the small model capacity, it integrates the attention-based feature fusion module and the multiple auxiliary losses enhancement strategy for better detecting subtle image changes and optimizing weights. Using transfer learning and five-fold cross-validation, Fast-MpoxNet achieves 94.26% Accuracy on the Mpox dataset. Notably, its recall for early-stage monkeypox achieves 93.65%. By adopting data augmentation, our model's Accuracy rises to 98.40% and attains a Practicality Score (A new metric for measuring model practicality in real-time diagnosis application) of 0.80. We also developed an application system named Mpox-AISM V2 for both personal computers and mobile phones. Mpox-AISM V2 features ultrafast responses, offline functionality, and easy deployment, enabling accurate and real-time diagnosis for both the public and individuals in various real-world settings, especially in populous settings during the outbreak. Our work could potentially mitigate future monkeypox outbreak and illuminate a fresh paradigm for developing real-time diagnostic tools in the healthcare field.
Ultrafast and Ultralight Network-Based Intelligent System for Real-time Diagnosis of Ear diseases in Any Devices
Aug 21, 2023



Abstract:Traditional ear disease diagnosis heavily depends on experienced specialists and specialized equipment, frequently resulting in misdiagnoses, treatment delays, and financial burdens for some patients. Utilizing deep learning models for efficient ear disease diagnosis has proven effective and affordable. However, existing research overlooked model inference speed and parameter size required for deployment. To tackle these challenges, we constructed a large-scale dataset comprising eight ear disease categories and normal ear canal samples from two hospitals. Inspired by ShuffleNetV2, we developed Best-EarNet, an ultrafast and ultralight network enabling real-time ear disease diagnosis. Best-EarNet incorporates the novel Local-Global Spatial Feature Fusion Module which can capture global and local spatial information simultaneously and guide the network to focus on crucial regions within feature maps at various levels, mitigating low accuracy issues. Moreover, our network uses multiple auxiliary classification heads for efficient parameter optimization. With 0.77M parameters, Best-EarNet achieves an average frames per second of 80 on CPU. Employing transfer learning and five-fold cross-validation with 22,581 images from Hospital-1, the model achieves an impressive 95.23% accuracy. External testing on 1,652 images from Hospital-2 validates its performance, yielding 92.14% accuracy. Compared to state-of-the-art networks, Best-EarNet establishes a new state-of-the-art (SOTA) in practical applications. Most importantly, we developed an intelligent diagnosis system called Ear Keeper, which can be deployed on common electronic devices. By manipulating a compact electronic otoscope, users can perform comprehensive scanning and diagnosis of the ear canal using real-time video. This study provides a novel paradigm for ear endoscopy and other medical endoscopic image recognition applications.
Mpox-AISM: AI-Mediated Super Monitoring for Forestalling Monkeypox Spread
Mar 17, 2023Abstract:The challenge on forestalling monkeypox (Mpox) spread is the timely, convenient and accurate diagnosis for earlystage infected individuals. Here, we propose a remote and realtime online visualization strategy, called "Super Monitoring" to construct a low cost, convenient, timely and unspecialized diagnosis of early-stage Mpox. Such AI-mediated "Super Monitoring" (Mpox-AISM) invokes a framework assembled by deep learning, data augmentation and self-supervised learning, as well as professionally classifies four subtypes according to dataset characteristics and evolution trend of Mpox and seven other types of dermatopathya with high similarity, hence these features together with reasonable program interface and threshold setting ensure that its Recall (Sensitivity) was beyond 95.9% and the specificity was almost 100%. As a result, with the help of cloud service on Internet and communication terminal, this strategy can be potentially utilized for the real-time detection of earlystage Mpox in various scenarios including entry-exit inspection in airport, family doctor, rural area in underdeveloped region and wild to effectively shorten the window period of Mpox spread.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge