Zekang Yang
PA-MIL: Phenotype-Aware Multiple Instance Learning Guided by Language Prompting and Genotype-to-Phenotype Relationships
Jan 30, 2026Abstract:Deep learning has been extensively researched in the analysis of pathology whole-slide images (WSIs). However, most existing methods are limited to providing prediction interpretability by locating the model's salient areas in a post-hoc manner, failing to offer more reliable and accountable explanations. In this work, we propose Phenotype-Aware Multiple Instance Learning (PA-MIL), a novel ante-hoc interpretable framework that identifies cancer-related phenotypes from WSIs and utilizes them for cancer subtyping. To facilitate PA-MIL in learning phenotype-aware features, we 1) construct a phenotype knowledge base containing cancer-related phenotypes and their associated genotypes. 2) utilize the morphological descriptions of phenotypes as language prompting to aggregate phenotype-related features. 3) devise the Genotype-to-Phenotype Neural Network (GP-NN) grounded in genotype-to-phenotype relationships, which provides multi-level guidance for PA-MIL. Experimental results on multiple datasets demonstrate that PA-MIL achieves competitive performance compared to existing MIL methods while offering improved interpretability. PA-MIL leverages phenotype saliency as evidence and, using a linear classifier, achieves competitive results compared to state-of-the-art methods. Additionally, we thoroughly analyze the genotype-phenotype relationships, as well as cohort-level and case-level interpretability, demonstrating the reliability and accountability of PA-MIL.
NADER: Neural Architecture Design via Multi-Agent Collaboration
Dec 26, 2024



Abstract:Designing effective neural architectures poses a significant challenge in deep learning. While Neural Architecture Search (NAS) automates the search for optimal architectures, existing methods are often constrained by predetermined search spaces and may miss critical neural architectures. In this paper, we introduce NADER (Neural Architecture Design via multi-agEnt collaboRation), a novel framework that formulates neural architecture design (NAD) as a LLM-based multi-agent collaboration problem. NADER employs a team of specialized agents to enhance a base architecture through iterative modification. Current LLM-based NAD methods typically operate independently, lacking the ability to learn from past experiences, which results in repeated mistakes and inefficient exploration. To address this issue, we propose the Reflector, which effectively learns from immediate feedback and long-term experiences. Additionally, unlike previous LLM-based methods that use code to represent neural architectures, we utilize a graph-based representation. This approach allows agents to focus on design aspects without being distracted by coding. We demonstrate the effectiveness of NADER in discovering high-performing architectures beyond predetermined search spaces through extensive experiments on benchmark tasks, showcasing its advantages over state-of-the-art methods. The codes will be released soon.
A Multimodal Object-level Contrast Learning Method for Cancer Survival Risk Prediction
Sep 03, 2024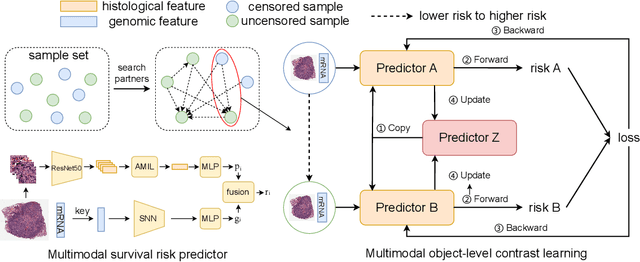
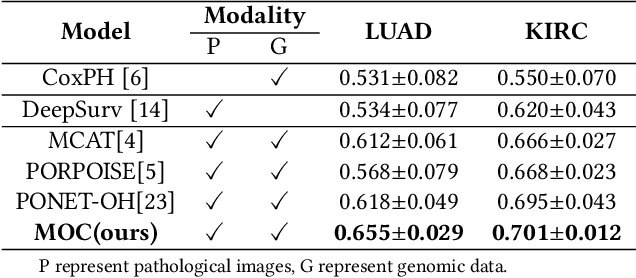
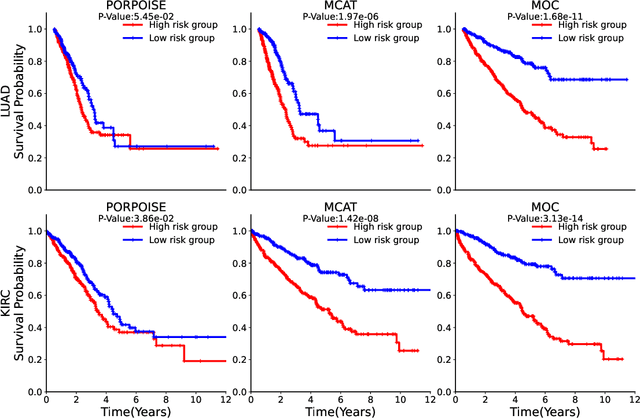
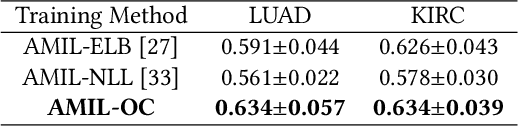
Abstract:Computer-aided cancer survival risk prediction plays an important role in the timely treatment of patients. This is a challenging weakly supervised ordinal regression task associated with multiple clinical factors involved such as pathological images, genomic data and etc. In this paper, we propose a new training method, multimodal object-level contrast learning, for cancer survival risk prediction. First, we construct contrast learning pairs based on the survival risk relationship among the samples in the training sample set. Then we introduce the object-level contrast learning method to train the survival risk predictor. We further extend it to the multimodal scenario by applying cross-modal constrast. Considering the heterogeneity of pathological images and genomics data, we construct a multimodal survival risk predictor employing attention-based and self-normalizing based nerural network respectively. Finally, the survival risk predictor trained by our proposed method outperforms state-of-the-art methods on two public multimodal cancer datasets for survival risk prediction.
SCMIL: Sparse Context-aware Multiple Instance Learning for Predicting Cancer Survival Probability Distribution in Whole Slide Images
Jun 30, 2024Abstract:Cancer survival prediction is a challenging task that involves analyzing of the tumor microenvironment within Whole Slide Image (WSI). Previous methods cannot effectively capture the intricate interaction features among instances within the local area of WSI. Moreover, existing methods for cancer survival prediction based on WSI often fail to provide better clinically meaningful predictions. To overcome these challenges, we propose a Sparse Context-aware Multiple Instance Learning (SCMIL) framework for predicting cancer survival probability distributions. SCMIL innovatively segments patches into various clusters based on their morphological features and spatial location information, subsequently leveraging sparse self-attention to discern the relationships between these patches with a context-aware perspective. Considering many patches are irrelevant to the task, we introduce a learnable patch filtering module called SoftFilter, which ensures that only interactions between task-relevant patches are considered. To enhance the clinical relevance of our prediction, we propose a register-based mixture density network to forecast the survival probability distribution for individual patients. We evaluate SCMIL on two public WSI datasets from the The Cancer Genome Atlas (TCGA) specifically focusing on lung adenocarcinom (LUAD) and kidney renal clear cell carcinoma (KIRC). Our experimental results indicate that SCMIL outperforms current state-of-the-art methods for survival prediction, offering more clinically meaningful and interpretable outcomes. Our code is accessible at https://github.com/yang-ze-kang/SCMIL.
AutoMMLab: Automatically Generating Deployable Models from Language Instructions for Computer Vision Tasks
Feb 23, 2024



Abstract:Automated machine learning (AutoML) is a collection of techniques designed to automate the machine learning development process. While traditional AutoML approaches have been successfully applied in several critical steps of model development (e.g. hyperparameter optimization), there lacks a AutoML system that automates the entire end-to-end model production workflow. To fill this blank, we present AutoMMLab, a general-purpose LLM-empowered AutoML system that follows user's language instructions to automate the whole model production workflow for computer vision tasks. The proposed AutoMMLab system effectively employs LLMs as the bridge to connect AutoML and OpenMMLab community, empowering non-expert individuals to easily build task-specific models via a user-friendly language interface. Specifically, we propose RU-LLaMA to understand users' request and schedule the whole pipeline, and propose a novel LLM-based hyperparameter optimizer called HPO-LLaMA to effectively search for the optimal hyperparameters. Experiments show that our AutoMMLab system is versatile and covers a wide range of mainstream tasks, including classification, detection, segmentation and keypoint estimation. We further develop a new benchmark, called LAMP, for studying key components in the end-to-end prompt-based model training pipeline. Code, model, and data will be released.
AIROGS: Artificial Intelligence for RObust Glaucoma Screening Challenge
Feb 10, 2023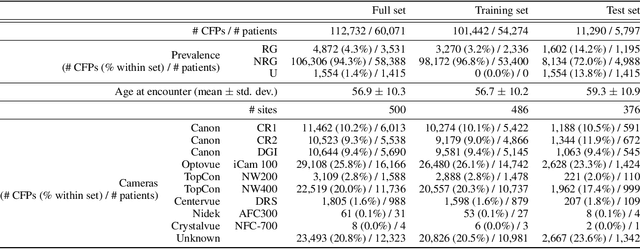
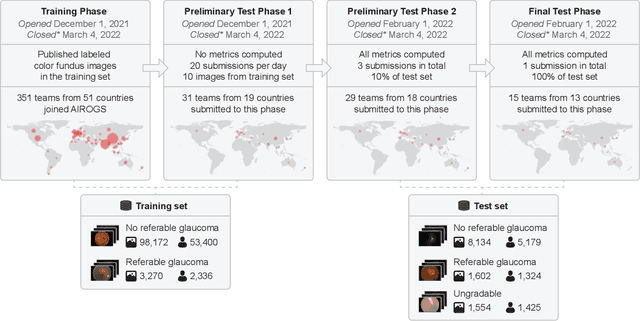
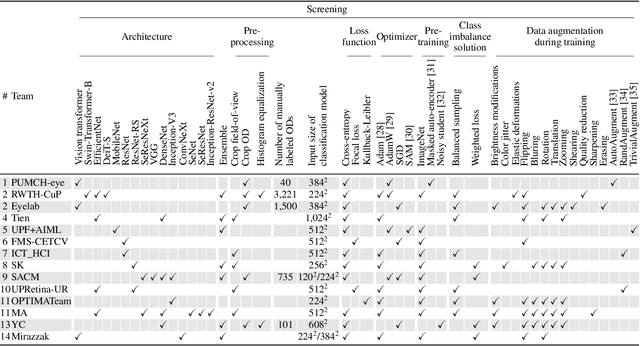
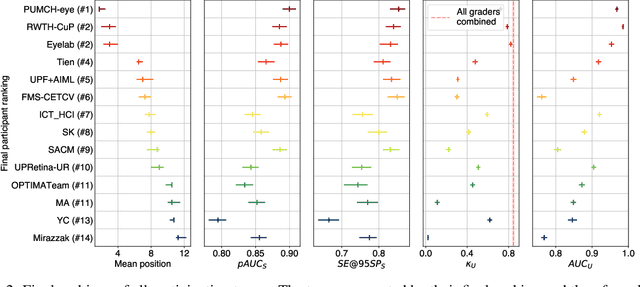
Abstract:The early detection of glaucoma is essential in preventing visual impairment. Artificial intelligence (AI) can be used to analyze color fundus photographs (CFPs) in a cost-effective manner, making glaucoma screening more accessible. While AI models for glaucoma screening from CFPs have shown promising results in laboratory settings, their performance decreases significantly in real-world scenarios due to the presence of out-of-distribution and low-quality images. To address this issue, we propose the Artificial Intelligence for Robust Glaucoma Screening (AIROGS) challenge. This challenge includes a large dataset of around 113,000 images from about 60,000 patients and 500 different screening centers, and encourages the development of algorithms that are robust to ungradable and unexpected input data. We evaluated solutions from 14 teams in this paper, and found that the best teams performed similarly to a set of 20 expert ophthalmologists and optometrists. The highest-scoring team achieved an area under the receiver operating characteristic curve of 0.99 (95% CI: 0.98-0.99) for detecting ungradable images on-the-fly. Additionally, many of the algorithms showed robust performance when tested on three other publicly available datasets. These results demonstrate the feasibility of robust AI-enabled glaucoma screening.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge