Yusheng Liao
AgentEHR: Advancing Autonomous Clinical Decision-Making via Retrospective Summarization
Jan 20, 2026Abstract:Large Language Models have demonstrated profound utility in the medical domain. However, their application to autonomous Electronic Health Records~(EHRs) navigation remains constrained by a reliance on curated inputs and simplified retrieval tasks. To bridge the gap between idealized experimental settings and realistic clinical environments, we present AgentEHR. This benchmark challenges agents to execute complex decision-making tasks, such as diagnosis and treatment planning, requiring long-range interactive reasoning directly within raw and high-noise databases. In tackling these tasks, we identify that existing summarization methods inevitably suffer from critical information loss and fractured reasoning continuity. To address this, we propose RetroSum, a novel framework that unifies a retrospective summarization mechanism with an evolving experience strategy. By dynamically re-evaluating interaction history, the retrospective mechanism prevents long-context information loss and ensures unbroken logical coherence. Additionally, the evolving strategy bridges the domain gap by retrieving accumulated experience from a memory bank. Extensive empirical evaluations demonstrate that RetroSum achieves performance gains of up to 29.16% over competitive baselines, while significantly decreasing total interaction errors by up to 92.3%.
Selecting Auxiliary Data via Neural Tangent Kernels for Low-Resource Domains
Nov 10, 2025Abstract:Large language models (LLMs) have achieved remarkable success across widespread tasks, yet their application in low-resource domains remains a significant challenge due to data scarcity and the high risk of overfitting. While in-domain data is limited, there exist vast amounts of similar general-domain data, and our initial findings reveal that they could potentially serve as auxiliary supervision for domain enhancement. This observation leads us to our central research question: \textbf{\textit{how to effectively select the most valuable auxiliary data to maximize domain-specific performance}}, particularly when traditional methods are inapplicable due to a lack of large in-domain data pools or validation sets. To address this, we propose \textbf{NTK-Selector}, a principled and efficient framework for selecting general-domain auxiliary data to enhance domain-specific performance via neural tangent kernels (NTK). Our method tackles two challenges of directly applying NTK to LLMs, theoretical assumptions and prohibitive computational cost, by empirically demonstrating a stable NTK-like behavior in LLMs during LoRA fine-tuning and proposing a Jacobian-free approximation method. Extensive experiments across four low-resource domains (medical, financial, legal, and psychological) demonstrate that NTK-Selector consistently improves downstream performance. Specifically, fine-tuning on 1,000 in-domain samples alone only yielded +0.8 points for Llama3-8B-Instruct and +0.9 points for Qwen3-8B. In contrast, enriching with 9,000 auxiliary samples selected by NTK-Selector led to substantial \textbf{gains of +8.7 and +5.1 points}, which corresponds to a \textbf{10.9x and 5.7x improvement} over the domain-only setting.
Bridging the Dynamic Perception Gap: Training-Free Draft Chain-of-Thought for Dynamic Multimodal Spatial Reasoning
May 22, 2025Abstract:While chains-of-thought (CoT) have advanced complex reasoning in multimodal large language models (MLLMs), existing methods remain confined to text or static visual domains, often faltering in dynamic spatial reasoning tasks. To bridge this gap, we present GRASSLAND, a novel maze navigation benchmark designed to evaluate dynamic spatial reasoning. Our experiments show that augmenting textual reasoning chains with dynamic visual drafts, overlaid on input images, significantly outperforms conventional approaches, offering new insights into spatial reasoning in evolving environments. To generalize this capability, we propose D2R (Dynamic Draft-Augmented Reasoning), a training-free framework that seamlessly integrates textual CoT with corresponding visual drafts into MLLMs. Extensive evaluations demonstrate that D2R consistently enhances performance across diverse tasks, establishing a robust baseline for dynamic spatial reasoning without requiring model fine-tuning. Project is open at https://github.com/Cratileo/D2R.
DSVD: Dynamic Self-Verify Decoding for Faithful Generation in Large Language Models
Mar 05, 2025Abstract:The reliability of large language models remains a critical challenge, particularly due to their susceptibility to hallucinations and factual inaccuracies during text generation. Existing solutions either underutilize models' self-correction with preemptive strategies or use costly post-hoc verification. To further explore the potential of real-time self-verification and correction, we present Dynamic Self-Verify Decoding (DSVD), a novel decoding framework that enhances generation reliability through real-time hallucination detection and efficient error correction. DSVD integrates two key components: (1) parallel self-verification architecture for continuous quality assessment, (2) dynamic rollback mechanism for targeted error recovery. Extensive experiments across five benchmarks demonstrate DSVD's effectiveness, achieving significant improvement in truthfulness (Quesetion-Answering) and factual accuracy (FActScore). Results show the DSVD can be further incorporated with existing faithful decoding methods to achieve stronger performance. Our work establishes that real-time self-verification during generation offers a viable path toward more trustworthy language models without sacrificing practical deployability.
MedS$^3$: Towards Medical Small Language Models with Self-Evolved Slow Thinking
Jan 21, 2025



Abstract:Medical language models (MLMs) have become pivotal in advancing medical natural language processing. However, prior models that rely on pre-training or supervised fine-tuning often exhibit low data efficiency and limited practicality in real-world clinical applications. While OpenAIs O1 highlights test-time scaling in mathematics, attempts to replicate this approach in medicine typically distill responses from GPT-series models to open-source models, focusing primarily on multiple-choice tasks. This strategy, though straightforward, neglects critical concerns like data privacy and realistic deployment in clinical settings. In this work, we present a deployable, small-scale medical language model, \mone, designed for long-chain reasoning in clinical tasks using a self-evolution paradigm. Starting with a seed dataset of around 8,000 instances spanning five domains and 16 datasets, we prompt a base policy model to perform Monte Carlo Tree Search (MCTS) to construct verifiable reasoning chains. Each reasoning step is assigned an evolution rollout value, allowing verified trajectories to train the policy model and the reward model. During inference, the policy model generates multiple responses, and the reward model selects the one with the highest reward score. Experiments on eleven evaluation datasets demonstrate that \mone outperforms prior open-source models by 2 points, with the addition of the reward model further boosting performance ($\sim$13 points), surpassing GPT-4o-mini. Code and data are available at \url{https://github.com/pixas/MedSSS}.
Towards Omni-RAG: Comprehensive Retrieval-Augmented Generation for Large Language Models in Medical Applications
Jan 05, 2025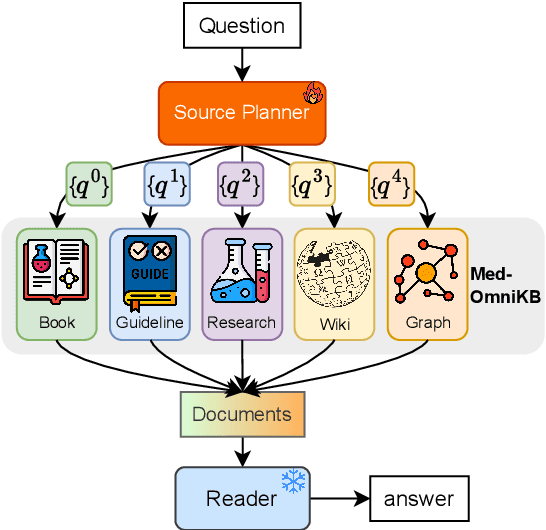
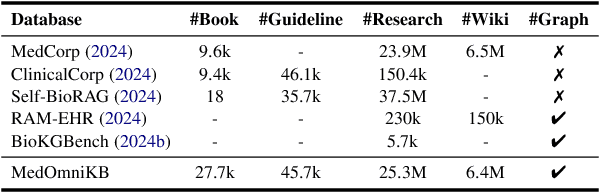
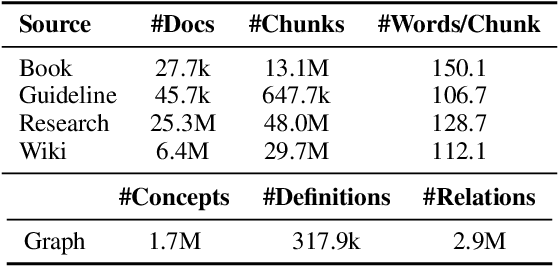
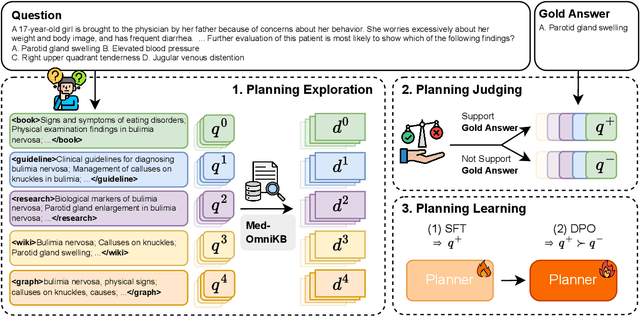
Abstract:Large language models (LLMs) hold promise for addressing healthcare challenges but often generate hallucinations due to limited integration of medical knowledge. Incorporating external medical knowledge is therefore critical, especially considering the breadth and complexity of medical content, which necessitates effective multi-source knowledge acquisition. We address this challenge by framing it as a source planning problem, where the task is to formulate context-appropriate queries tailored to the attributes of diverse knowledge sources. Existing approaches either overlook source planning or fail to achieve it effectively due to misalignment between the model's expectation of the sources and their actual content. To bridge this gap, we present MedOmniKB, a comprehensive repository comprising multigenre and multi-structured medical knowledge sources. Leveraging these sources, we propose the Source Planning Optimisation (SPO) method, which enhances multi-source utilisation through explicit planning optimisation. Our approach involves enabling an expert model to explore and evaluate potential plans while training a smaller model to learn source alignment using positive and negative planning samples. Experimental results demonstrate that our method substantially improves multi-source planning performance, enabling the optimised small model to achieve state-of-the-art results in leveraging diverse medical knowledge sources.
Drawing the Line: Enhancing Trustworthiness of MLLMs Through the Power of Refusal
Dec 15, 2024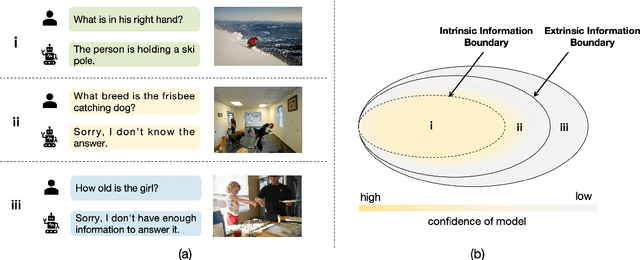
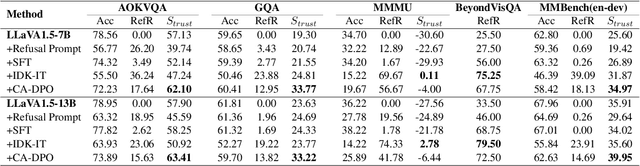
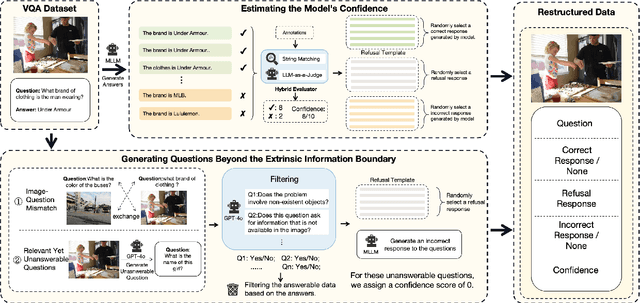
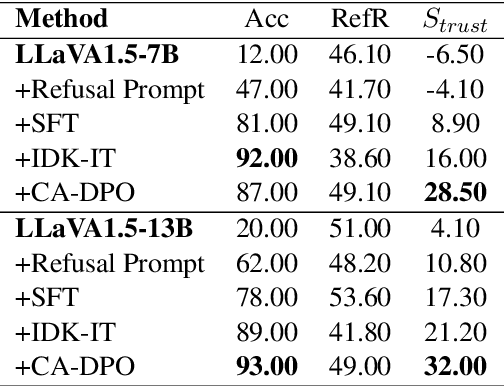
Abstract:Multimodal large language models (MLLMs) excel at multimodal perception and understanding, yet their tendency to generate hallucinated or inaccurate responses undermines their trustworthiness. Existing methods have largely overlooked the importance of refusal responses as a means of enhancing MLLMs reliability. To bridge this gap, we present the Information Boundary-aware Learning Framework (InBoL), a novel approach that empowers MLLMs to refuse to answer user queries when encountering insufficient information. To the best of our knowledge, InBoL is the first framework that systematically defines the conditions under which refusal is appropriate for MLLMs using the concept of information boundaries proposed in our paper. This framework introduces a comprehensive data generation pipeline and tailored training strategies to improve the model's ability to deliver appropriate refusal responses. To evaluate the trustworthiness of MLLMs, we further propose a user-centric alignment goal along with corresponding metrics. Experimental results demonstrate a significant improvement in refusal accuracy without noticeably compromising the model's helpfulness, establishing InBoL as a pivotal advancement in building more trustworthy MLLMs.
ReflecTool: Towards Reflection-Aware Tool-Augmented Clinical Agents
Oct 23, 2024Abstract:Large Language Models (LLMs) have shown promising potential in the medical domain, assisting with tasks like clinical note generation and patient communication. However, current LLMs are limited to text-based communication, hindering their ability to interact with diverse forms of information in clinical environments. Despite clinical agents succeeding in diverse signal interaction, they are oriented to a single clinical scenario and hence fail for broader applications. To evaluate clinical agents holistically, we propose ClinicalAgent Bench~(CAB), a comprehensive medical agent benchmark consisting of 18 tasks across five key realistic clinical dimensions. Building on this, we introduce ReflecTool, a novel framework that excels at utilizing domain-specific tools within two stages. The first optimization stage progressively enlarges a long-term memory by saving successful solving processes and tool-wise experience of agents in a tiny pre-defined training set. In the following inference stage, ReflecTool can search for supportive successful demonstrations from already built long-term memory to guide the tool selection strategy, and a verifier improves the tool usage according to the tool-wise experience with two verification methods--iterative refinement and candidate selection. Extensive experiments on ClinicalAgent Benchmark demonstrate that ReflecTool surpasses the pure LLMs with more than 10 points and the well-established agent-based methods with 3 points, highlighting its adaptability and effectiveness in solving complex clinical tasks.
Med-PMC: Medical Personalized Multi-modal Consultation with a Proactive Ask-First-Observe-Next Paradigm
Aug 16, 2024



Abstract:The application of the Multi-modal Large Language Models (MLLMs) in medical clinical scenarios remains underexplored. Previous benchmarks only focus on the capacity of the MLLMs in medical visual question-answering (VQA) or report generation and fail to assess the performance of the MLLMs on complex clinical multi-modal tasks. In this paper, we propose a novel Medical Personalized Multi-modal Consultation (Med-PMC) paradigm to evaluate the clinical capacity of the MLLMs. Med-PMC builds a simulated clinical environment where the MLLMs are required to interact with a patient simulator to complete the multi-modal information-gathering and decision-making task. Specifically, the patient simulator is decorated with personalized actors to simulate diverse patients in real scenarios. We conduct extensive experiments to access 12 types of MLLMs, providing a comprehensive view of the MLLMs' clinical performance. We found that current MLLMs fail to gather multimodal information and show potential bias in the decision-making task when consulted with the personalized patient simulators. Further analysis demonstrates the effectiveness of Med-PMC, showing the potential to guide the development of robust and reliable clinical MLLMs. Code and data are available at https://github.com/LiuHC0428/Med-PMC.
Decoding Linguistic Representations of Human Brain
Jul 30, 2024



Abstract:Language, as an information medium created by advanced organisms, has always been a concern of neuroscience regarding how it is represented in the brain. Decoding linguistic representations in the evoked brain has shown groundbreaking achievements, thanks to the rapid improvement of neuroimaging, medical technology, life sciences and artificial intelligence. In this work, we present a taxonomy of brain-to-language decoding of both textual and speech formats. This work integrates two types of research: neuroscience focusing on language understanding and deep learning-based brain decoding. Generating discernible language information from brain activity could not only help those with limited articulation, especially amyotrophic lateral sclerosis (ALS) patients but also open up a new way for the next generation's brain-computer interface (BCI). This article will help brain scientists and deep-learning researchers to gain a bird's eye view of fine-grained language perception, and thus facilitate their further investigation and research of neural process and language decoding.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge