Yuli Wu
Eye on the Target: Eye Tracking Meets Rodent Tracking
Mar 13, 2025Abstract:Analyzing animal behavior from video recordings is crucial for scientific research, yet manual annotation remains labor-intensive and prone to subjectivity. Efficient segmentation methods are needed to automate this process while maintaining high accuracy. In this work, we propose a novel pipeline that utilizes eye-tracking data from Aria glasses to generate prompt points, which are then used to produce segmentation masks via a fast zero-shot segmentation model. Additionally, we apply post-processing to refine the prompts, leading to improved segmentation quality. Through our approach, we demonstrate that combining eye-tracking-based annotation with smart prompt refinement can enhance segmentation accuracy, achieving an improvement of 70.6% from 38.8 to 66.2 in the Jaccard Index for segmentation results in the rats dataset.
CellStyle: Improved Zero-Shot Cell Segmentation via Style Transfer
Mar 11, 2025Abstract:Cell microscopy data are abundant; however, corresponding segmentation annotations remain scarce. Moreover, variations in cell types, imaging devices, and staining techniques introduce significant domain gaps between datasets. As a result, even large, pretrained segmentation models trained on diverse datasets (source datasets) struggle to generalize to unseen datasets (target datasets). To overcome this generalization problem, we propose CellStyle, which improves the segmentation quality of such models without requiring labels for the target dataset, thereby enabling zero-shot adaptation. CellStyle transfers the attributes of an unannotated target dataset, such as texture, color, and noise, to the annotated source dataset. This transfer is performed while preserving the cell shapes of the source images, ensuring that the existing source annotations can still be used while maintaining the visual characteristics of the target dataset. The styled synthetic images with the existing annotations enable the finetuning of a generalist segmentation model for application to the unannotated target data. We demonstrate that CellStyle significantly improves zero-shot cell segmentation performance across diverse datasets by finetuning multiple segmentation models on the style-transferred data. The code will be made publicly available.
A Pragmatic Note on Evaluating Generative Models with Fréchet Inception Distance for Retinal Image Synthesis
Feb 26, 2025Abstract:Fr\'echet Inception Distance (FID), computed with an ImageNet pretrained Inception-v3 network, is widely used as a state-of-the-art evaluation metric for generative models. It assumes that feature vectors from Inception-v3 follow a multivariate Gaussian distribution and calculates the 2-Wasserstein distance based on their means and covariances. While FID effectively measures how closely synthetic data match real data in many image synthesis tasks, the primary goal in biomedical generative models is often to enrich training datasets ideally with corresponding annotations. For this purpose, the gold standard for evaluating generative models is to incorporate synthetic data into downstream task training, such as classification and segmentation, to pragmatically assess its performance. In this paper, we examine cases from retinal imaging modalities, including color fundus photography and optical coherence tomography, where FID and its related metrics misalign with task-specific evaluation goals in classification and segmentation. We highlight the limitations of using various metrics, represented by FID and its variants, as evaluation criteria for these applications and address their potential caveats in broader biomedical imaging modalities and downstream tasks.
No Free Lunch in Annotation either: An objective evaluation of foundation models for streamlining annotation in animal tracking
Feb 06, 2025Abstract:We analyze the capabilities of foundation models addressing the tedious task of generating annotations for animal tracking. Annotating a large amount of data is vital and can be a make-or-break factor for the robustness of a tracking model. Robustness is particularly crucial in animal tracking, as accurate tracking over long time horizons is essential for capturing the behavior of animals. However, generating additional annotations using foundation models can be counterproductive, as the quality of the annotations is just as important. Poorly annotated data can introduce noise and inaccuracies, ultimately compromising the performance and accuracy of the trained model. Over-reliance on automated annotations without ensuring precision can lead to diminished results, making careful oversight and quality control essential in the annotation process. Ultimately, we demonstrate that a thoughtful combination of automated annotations and manually annotated data is a valuable strategy, yielding an IDF1 score of 80.8 against blind usage of SAM2 video with an IDF1 score of 65.6.
Cascaded Diffusion Models for 2D and 3D Microscopy Image Synthesis to Enhance Cell Segmentation
Nov 19, 2024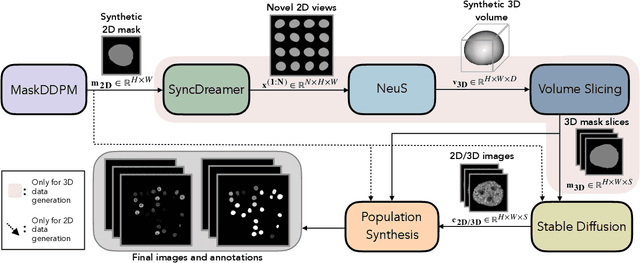
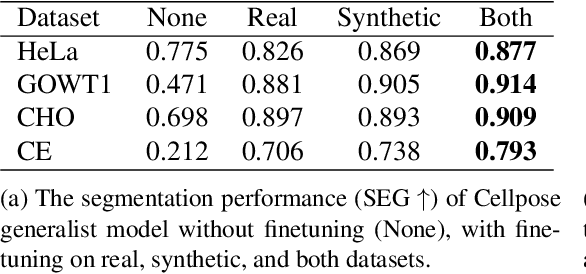
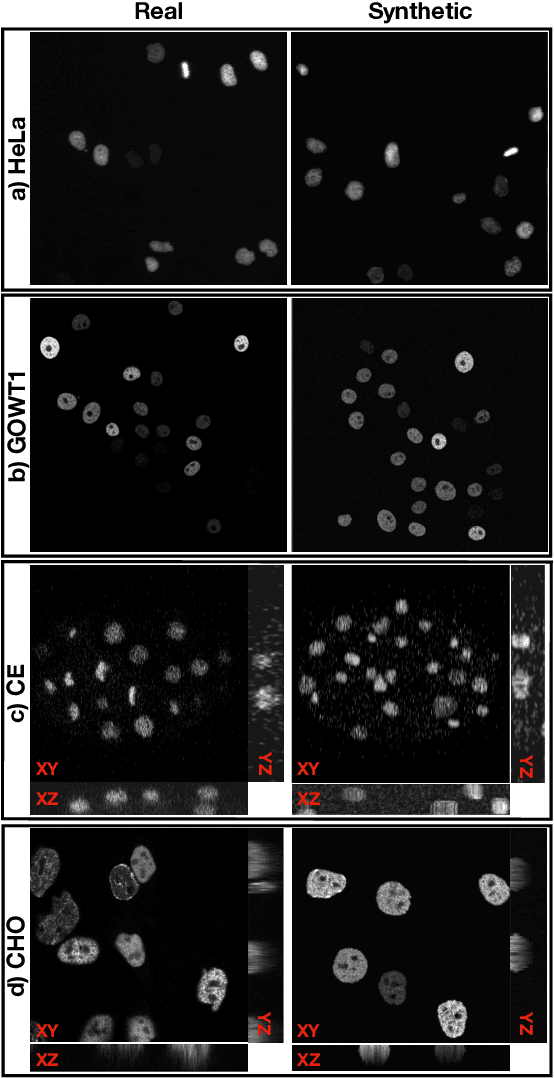
Abstract:Automated cell segmentation in microscopy images is essential for biomedical research, yet conventional methods are labor-intensive and prone to error. While deep learning-based approaches have proven effective, they often require large annotated datasets, which are scarce due to the challenges of manual annotation. To overcome this, we propose a novel framework for synthesizing densely annotated 2D and 3D cell microscopy images using cascaded diffusion models. Our method synthesizes 2D and 3D cell masks from sparse 2D annotations using multi-level diffusion models and NeuS, a 3D surface reconstruction approach. Following that, a pretrained 2D Stable Diffusion model is finetuned to generate realistic cell textures and the final outputs are combined to form cell populations. We show that training a segmentation model with a combination of our synthetic data and real data improves cell segmentation performance by up to 9\% across multiple datasets. Additionally, the FID scores indicate that the synthetic data closely resembles real data. The code for our proposed approach will be available at https://github.com/ruveydayilmaz0/cascaded_diffusion.
Visual Fixation-Based Retinal Prosthetic Simulation
Oct 15, 2024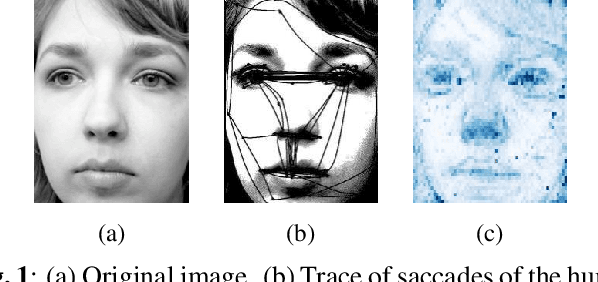
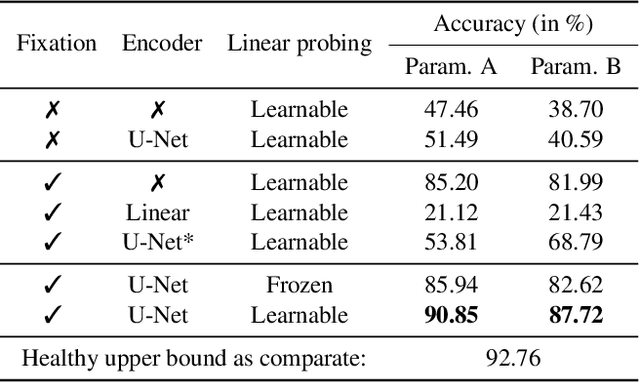
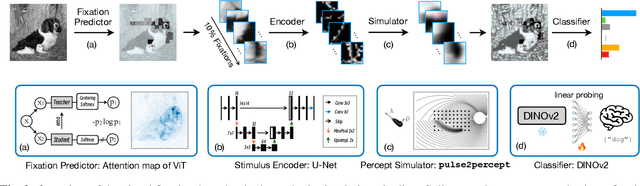
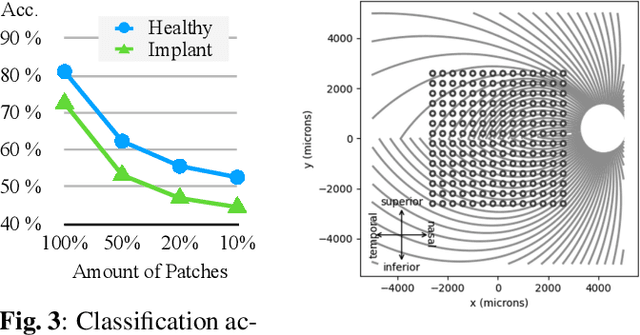
Abstract:This study proposes a retinal prosthetic simulation framework driven by visual fixations, inspired by the saccade mechanism, and assesses performance improvements through end-to-end optimization in a classification task. Salient patches are predicted from input images using the self-attention map of a vision transformer to mimic visual fixations. These patches are then encoded by a trainable U-Net and simulated using the pulse2percept framework to predict visual percepts. By incorporating a learnable encoder, we aim to optimize the visual information transmitted to the retinal implant, addressing both the limited resolution of the electrode array and the distortion between the input stimuli and resulting phosphenes. The predicted percepts are evaluated using the self-supervised DINOv2 foundation model, with an optional learnable linear layer for classification accuracy. On a subset of the ImageNet validation set, the fixation-based framework achieves a classification accuracy of 87.72%, using computational parameters based on a real subject's physiological data, significantly outperforming the downsampling-based accuracy of 40.59% and approaching the healthy upper bound of 92.76%. Our approach shows promising potential for producing more semantically understandable percepts with the limited resolution available in retinal prosthetics.
Optimizing Retinal Prosthetic Stimuli with Conditional Invertible Neural Networks
Mar 07, 2024Abstract:Implantable retinal prostheses offer a promising solution to restore partial vision by circumventing damaged photoreceptor cells in the retina and directly stimulating the remaining functional retinal cells. However, the information transmission between the camera and retinal cells is often limited by the low resolution of the electrode array and the lack of specificity for different ganglion cell types, resulting in suboptimal stimulations. In this work, we propose to utilize normalizing flow-based conditional invertible neural networks to optimize retinal implant stimulation in an unsupervised manner. The invertibility of these networks allows us to use them as a surrogate for the computational model of the visual system, while also encoding input camera signals into optimized electrical stimuli on the electrode array. Compared to other methods, such as trivial downsampling, linear models, and feed-forward convolutional neural networks, the flow-based invertible neural network and its conditional extension yield better visual reconstruction qualities w.r.t. various metrics using a physiologically validated simulation tool.
Loss Functions in the Era of Semantic Segmentation: A Survey and Outlook
Dec 08, 2023



Abstract:Semantic image segmentation, the process of classifying each pixel in an image into a particular class, plays an important role in many visual understanding systems. As the predominant criterion for evaluating the performance of statistical models, loss functions are crucial for shaping the development of deep learning-based segmentation algorithms and improving their overall performance. To aid researchers in identifying the optimal loss function for their particular application, this survey provides a comprehensive and unified review of $25$ loss functions utilized in image segmentation. We provide a novel taxonomy and thorough review of how these loss functions are customized and leveraged in image segmentation, with a systematic categorization emphasizing their significant features and applications. Furthermore, to evaluate the efficacy of these methods in real-world scenarios, we propose unbiased evaluations of some distinct and renowned loss functions on established medical and natural image datasets. We conclude this review by identifying current challenges and unveiling future research opportunities. Finally, we have compiled the reviewed studies that have open-source implementations on our GitHub page.
Retinal OCT Synthesis with Denoising Diffusion Probabilistic Models for Layer Segmentation
Nov 09, 2023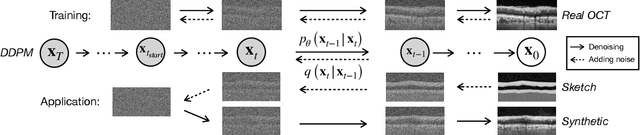
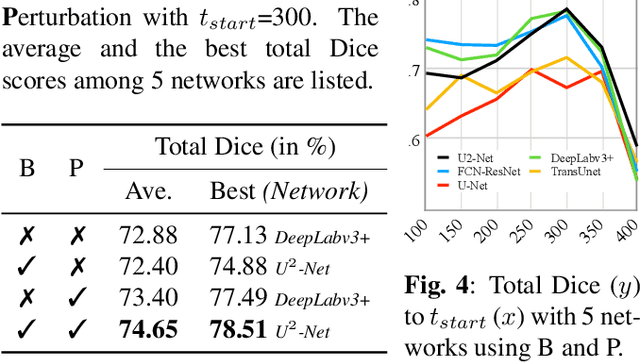
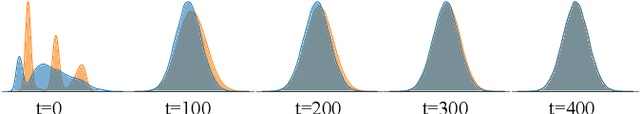
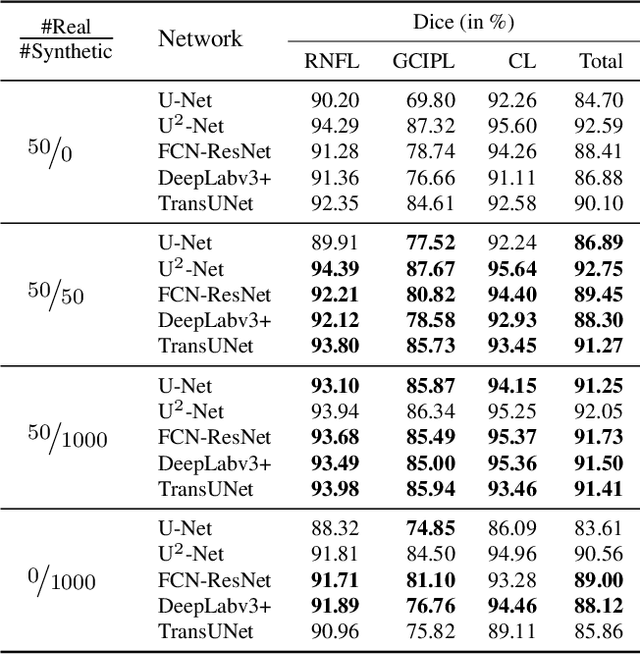
Abstract:Modern biomedical image analysis using deep learning often encounters the challenge of limited annotated data. To overcome this issue, deep generative models can be employed to synthesize realistic biomedical images. In this regard, we propose an image synthesis method that utilizes denoising diffusion probabilistic models (DDPMs) to automatically generate retinal optical coherence tomography (OCT) images. By providing rough layer sketches, the trained DDPMs can generate realistic circumpapillary OCT images. We further find that more accurate pseudo labels can be obtained through knowledge adaptation, which greatly benefits the segmentation task. Through this, we observe a consistent improvement in layer segmentation accuracy, which is validated using various neural networks. Furthermore, we have discovered that a layer segmentation model trained solely with synthesized images can achieve comparable results to a model trained exclusively with real images. These findings demonstrate the promising potential of DDPMs in reducing the need for manual annotations of retinal OCT images.
Semi-supervised Instance Segmentation with a Learned Shape Prior
Sep 09, 2023Abstract:To date, most instance segmentation approaches are based on supervised learning that requires a considerable amount of annotated object contours as training ground truth. Here, we propose a framework that searches for the target object based on a shape prior. The shape prior model is learned with a variational autoencoder that requires only a very limited amount of training data: In our experiments, a few dozens of object shape patches from the target dataset, as well as purely synthetic shapes, were sufficient to achieve results en par with supervised methods with full access to training data on two out of three cell segmentation datasets. Our method with a synthetic shape prior was superior to pre-trained supervised models with access to limited domain-specific training data on all three datasets. Since the learning of prior models requires shape patches, whether real or synthetic data, we call this framework semi-supervised learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge